This lecture can be viewed on YouTube site: Life Scinence Lectures for you
https://www.youtube.com/watch?v=lFGCRIQ9LPs
In this lecture, we will discuss the mechanism of transcriptional regulation of Tryptophan operon. This lecture contains 9 figures.
- The mechanism of transcriptional regulation in the tryptophan operon
- The mechanism of transcription attenuation by the attenuator
- Inhibition of translation initiation using structural changes in riboswitches and RNA aptamers
The function of the riboswitch in the thiMD operon of E. coli
Key words: tryptophan operon, riboswitch, RNA aptamer, Attenuator, Ribosome mediated attenuator, thiMD operon, E. coli
1. When Tryptophan concentration is low
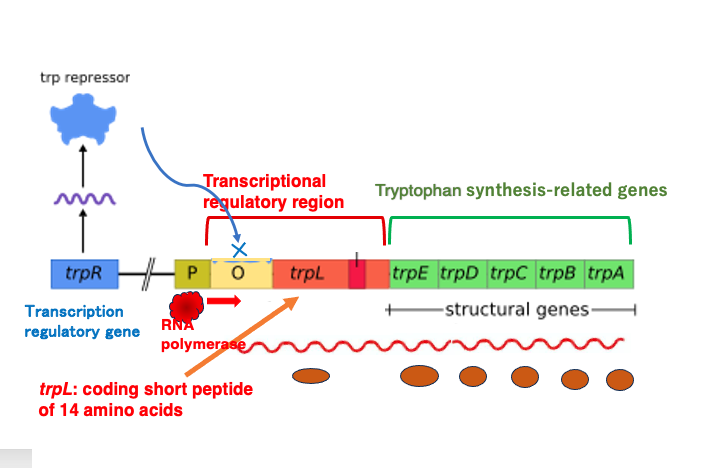
In E. coli, there is a Tryptophan operon responsible for synthesizing the amino acid Tryptophan. The Tryptophan operon contains five genes, from trpE to trpA, which encode the enzymes necessary for Tryptophan synthesis. In the transcriptional regulatory region, there is an open reading frame called trpL that produces a short peptide of 14 amino acids.
Upstream of this Tryptophan operon, there is a regulatory gene called trpR, which is constitutively expressed. Its product functions as a transcriptional repressor. When Tryptophan is present at low concentrations in the cytoplasm, most of these repressor proteins are not bound to Tryptophan and maintain their original conformation. In this state, they cannot bind to the DNA operator sequence.
As a result, RNA polymerase bound to the promoter can proceed with transcription normally, and the genes necessary for Tryptophan synthesis are transcribed. Consequently, the concentration of Tryptophan in the cytoplasm will eventually increase.
2. When Tryptophan concentration is high
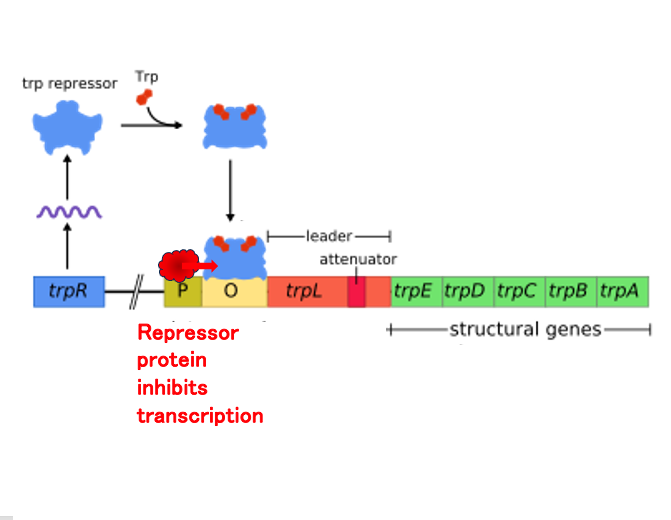
On the other hand, when Tryptophan is present in high concentrations in cytoplasm, many of the repressor proteins bind to Tryptophan, causing a change in the three-dimensional structure.
This repressor with the altered structure can attach to the operator, so even if RNA polymerase binds to the promoter and initiates transcription, it cannot transcribe the group of genes necessary for Tryptophan synthesis that are located downstream.
As a result, the concentration of Tryptophan will eventually decrease.
3. Ribosome-mediated attenuation
In addition to this Tryptophan concentration adjustment system, there is a system called ribosome-mediated attenuation that regulates the transcription amount of the operon.
This system takes advantage of the fact that in prokaryotes, translation can occur on mRNA that is still in the process of being transcribed. The term ‘attenuation’ refers to reduction or weakening.
Therefore, ribosome-mediated attenuation refers to the reduction of the transcripts of the Tryptophan operon, mediated by ribosomes.
4. Leader sequence region
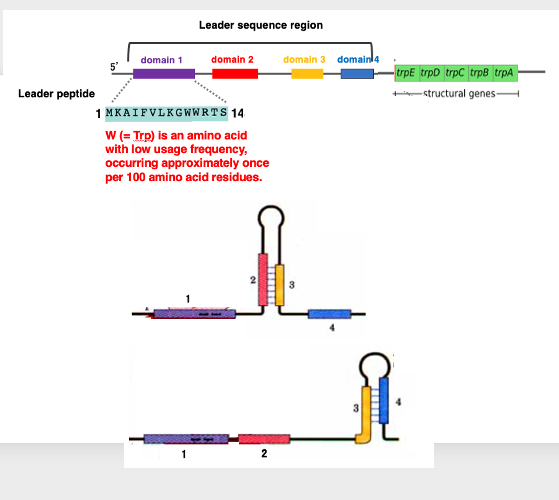
To explain this mechanism, let’s examine the Leader sequence region of the Tryptophan operon in detail.
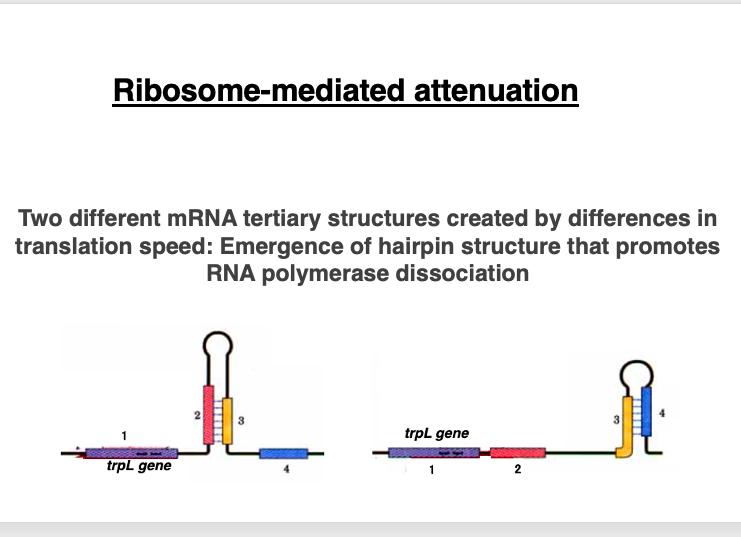
The Leader sequence region consists of four domains. The first domain encodes a short peptide composed of 14 amino acids. Interestingly, these domains can form interesting three-dimensional structures due to complementary sequences within each domain. Domains 2 and 3 can form a hairpin structure as shown on the left side of the figure below. Similarly, domains 3 and 4 can form another hairpin structure as shown on the right side of the figure.
This means that domain 3 has sequences capable of forming hairpin structures with both domain 2 and domain 4. This Leader sequence region is located upstream of the trpE to trpA genes, which are responsible for Tryptophan biosynthesis.
The 14-amino acid peptide encoded in Domain 1 has a very distinctive sequence, containing two Tryptophan residues. Tryptophan is an infrequently used amino acid, typically occurring only once in about 100 amino acid residues. The fact that it appears twice in such a short 14-amino acid sequence is remarkably characteristic, considering its low frequency of use.
5. Transcription and Translation: when Trp concentration is low
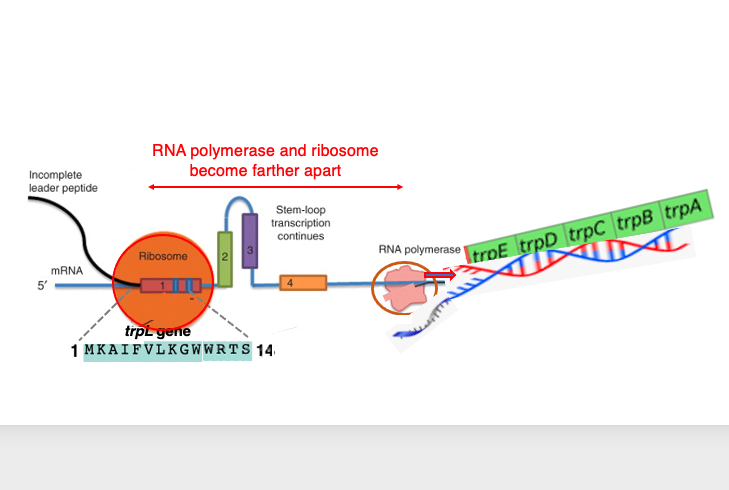
Let’s consider the relationship between transcription and translation when tryptophan levels are low. Since the concentration of tryptophan is low, the repressor is not attached to the operator sequence, allowing RNA polymerase to pass through the operator sequence and proceed with mRNA synthesis smoothly.
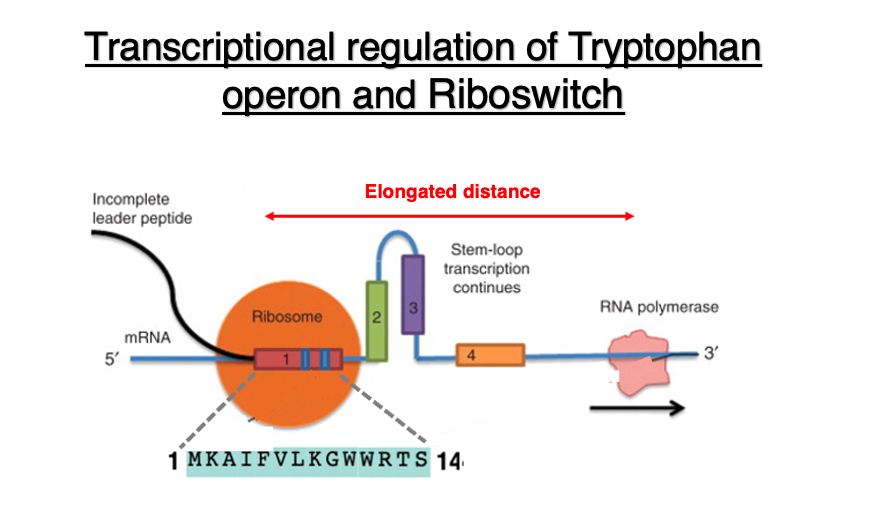
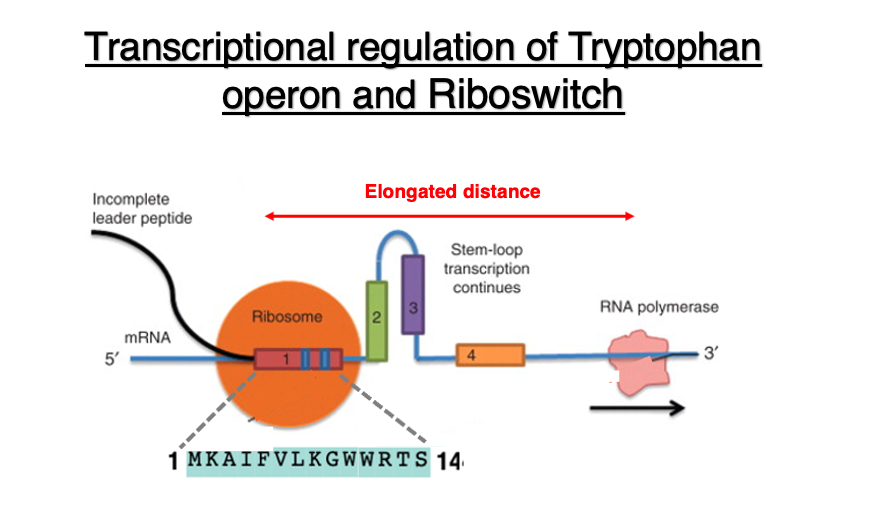
As shown in the diagram, the ribosome is translating the 14-peptide segment encoded in domain 1. Due to the low concentration of tryptophan in the cytoplasm, tryptophan-charged tRNA does not readily enter the ribosome when it tries to translate the tryptophan codon.
As a result, the ribosome is stalled at the tryptophan codon. Meanwhile, RNA polymerase continues transcription, leading to an increased distance. As a result, a hairpin is formed between Domain 2 and domain 3. This hairpin structure is located far from the RNA polymerase, so it does not affect the progress of transcription.
6. Transcription and Translation: when Trp concentration is high
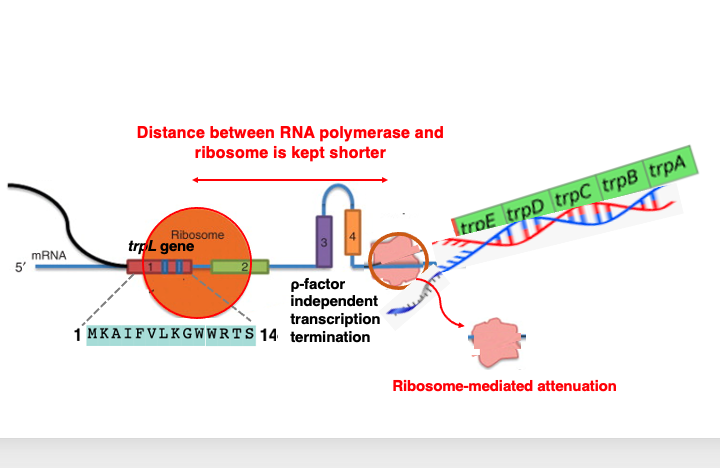
Regarding the relationship between transcription and translation when tryptophan concentration is high: In this case, it doesn’t take much time to translate the two tryptophan codons in the leader peptide region.
Consequently, when examining the relative positions of the ribosome and RNA polymerase, the distance between these two apparatuses is relatively short. This leads to the formation of a hairpin structure in domains 3 and 4. The hairpin structure positioned immediately behind the RNA polymerase can sometimes function as a ρ-factor independent transcription termination signal.
This causes the RNA polymerase to detach from the DNA sequence mid-transcription, resulting in premature termination of transcription. This phenomenon is called transcriptional attenuation, which leads to a decrease in transcription. This mechanism is referred to as ribosome-mediated attenuation.
In this way, when the concentration of tryptophan is high, the repressor undergoes a conformational change and attaches to the operator, thereby stopping the movement of RNA polymerase. Additionally, it can also halt the transcription process of mRNA that has already started to be synthesized. In bacteria, under conditions of high tryptophan concentration, the tryptophan operon transcription is suppressed using these two mechanisms.
7. Translation Regulation by Riboswitch
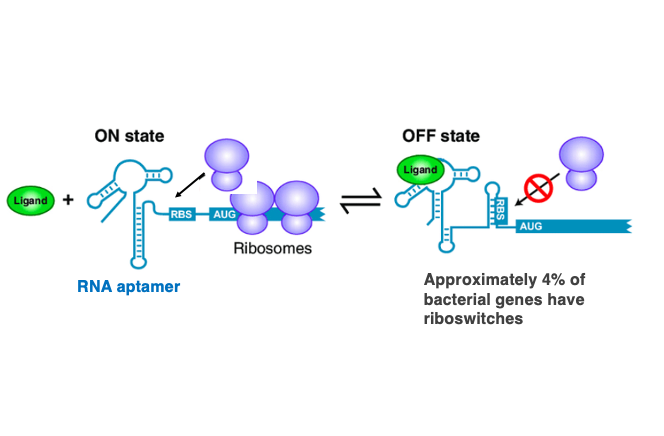
We’ve discussed how mRNA secondary structures can impede transcription continuation. Related to this topic, I’d like to explain about riboswitches. A riboswitch refers to a feedback mechanism that uses short RNA sequences called RNA aptamers to detect the concentration of small metabolites in the cell and suppress the expression of genes located downstream of the RNA aptamer.
This is an illustration of a riboswitch. Multiple hairpin structures are arranged at the 5′-end of the mRNA, forming a complex three-dimensional structure. When the 5′-end of the mRNA has the structure shown on the left, the Ribosome Binding Site (RBS) is exposed, allowing the ribosome to start translating the mRNA. However, when the low-molecular-weight product synthesized by the transcription of this gene accumulates to a high concentration, this substance ( Ligand ) attaches to the riboswitch.
This causes a change in the three-dimensional structure of the RNA at the 5′-end, making RBS part of a hairpin sequence. In this state, the efficiency of the ribosome attaching to the mRNA decreases. This is the mechanism of translation ON and OFF by the riboswitch. Since the riboswitch sequence specifically adsorbs certain low-molecular-weight metabolites, it can be considered a type of RNA aptamer. Riboswitches are present in approximately 4% of bacterial genes.
8. E. coli thiM riboswitch
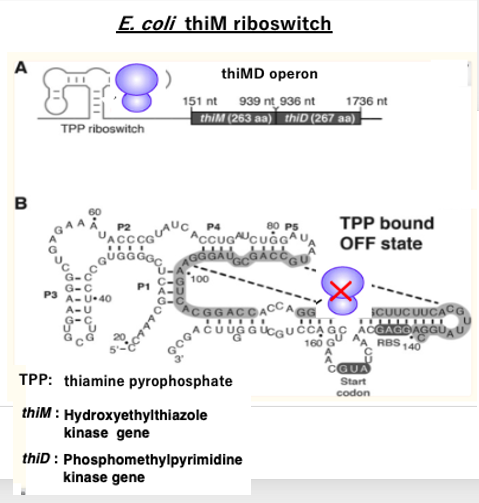
I will explain a specific example of a riboswitch in E. coli. In E. coli, there are two kinase genes, thiM and thiD, involved in the production of thiamine pyrophosphate (TPP).
These two kinase genes form an operon called the thiMD operon. The mRNA transcribed from this operon has an RNA aptamer structure added to its 5′ end, which functions as a riboswitch. When the concentration of the product TPP is low, the riboswitch assumes the structure shown in Figure A, and the translation of the mRNA proceeds normally. However, when the TPP concentration becomes high, TPP binds to this riboswitch, causing it to change into the three-dimensional structure shown in Figure B.
In this structure, the ribosome binding site and the translation initiation AUG codon are embedded within a hairpin structure, thereby inhibiting the initiation of translation. In this way, when the TPP concentration is high, the translation initiation of the messenger RNA is inhibited, thus suppressing the production of TPP.
9. Binding of a ligand changes the aptamer structure
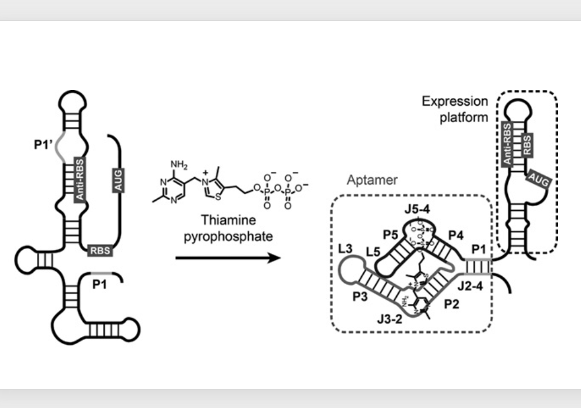
Let me explain using a different diagram. When thiamine pyrophosphate (TPP) is not attached, the Ribosome Binding Site (RBS) and the translation start codon are exposed, allowing translation to begin normally.
However, when the concentration of TPP increases, TPP binds to this RNA aptamer at the 5′ end of the mRNA, changing its three-dimensional structure. As the structure changes, the Ribosome Binding Site and translation start codon become buried within a hairpin structure, preventing normal translation initiation.
This riboswitch mechanism is used not only in bacteria but also in eukaryotes. With this, I conclude the explanation of the Tryptophan operon and riboswitch.
コメント