This lecture can be viewed as a video on YouTube site: Life Scinence Lectures for you
This lecture can be viewed as a video on YouTube site: Life Scinence Lectures for you
https://www.youtube.com/watch?v=qpJqx92o_jk&list=PL_B52Q_vHW1anGB1o8Fs-VL7UsUrhI1w4&index=5&t=220s
1. Contents of this lecture
This video contains the following and 32 figures.
1. Basic structure of viral particles
2. Classification of viruses based on how viruses multiply and their genomic nucleic acids
3. Carcinogenesis by DNA viral infection: Cervical cancer caused by papillomavirus infection
4. Infection and pathogenesis of viruses amplified by RNA dependent RNA polymerase: the case of Coronaviruses
5. Infection and pathogenesis of retroviruses: Acquired immunodeficiency syndrome due to AIDS virus (HIV) infection
6. Retroviruses carrying oncogenes in their genomes: Mechanism of sarcoma development caused by Rous sarcoma virus infection
Key words: Capsid, Envelope, Endocytosis, Plus chain, Minus chain, RNA dependent RNA polymerase, Reverse transcriptase, phage, bacteriophage, Cervical cancer, HPV, Corona virus, SARS, covid-19, spike protein, MERS, bat, natural host, alveolar epithelial cell, mutation notation, remdesivir, HIV, AIDS virus, acquired immune deficiency syndrome latency, LTR, long terminal repeat, CD4 receptor, gag, pol, env,p rovirus, lymphocyte, candidiasis, oncogene, v-src, c-src, proto-oncogene
2. Appearances of various viruses
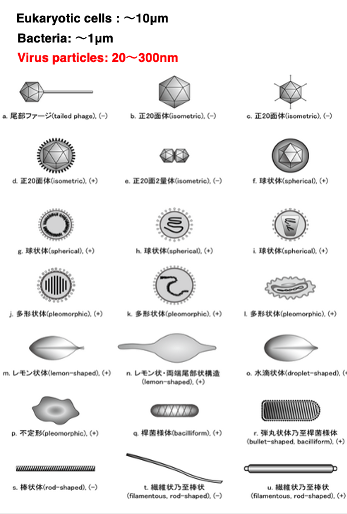
Here, I am showing the appearances of various viruses. As you can see, the appearances of viruses are quite diverse.
First, regarding their size, eukaryotic cells are about 10 micrometers, bacteria are around 1 μm, while viruses range from 20 to 300 nm
. 300 nanometers is equivalent to 0.3 μm. The limit of visibility for optical microscopes is 200 nm, so most viruses cannot be observed with optical microscopes. An electron microscope is necessary.
2. Basic structure of viruses
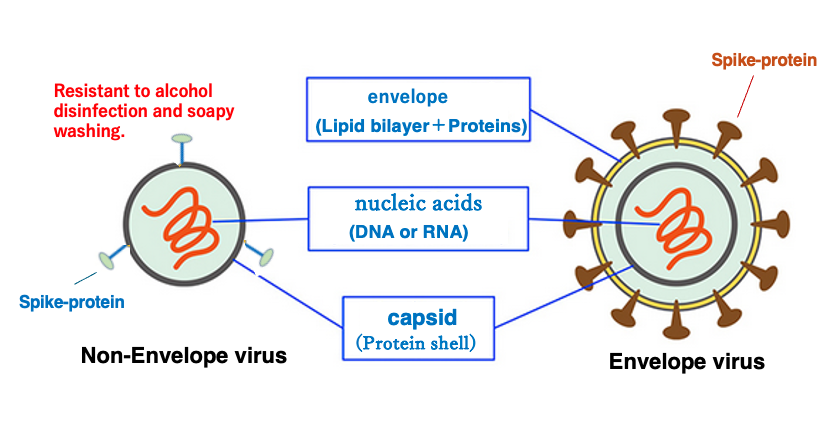
Here is the basic structure of viruses. First, viruses are classified into two categories.
One is the envelope virus, which has an envelope structure consisting of lipid bilayer at the outermost. Second is the non-envelope virus, which does not have an envelope structure. In the envelope lipid bilayer various proteins are embedded.
Since the envelope is a lipid bilayer, it is easily destroyed by alcohol and soap. On the other hand, the capsid is a protein shell, so it shows some resistance to alcohol and detergents. Thus, it follows that non-envelope viruses are more difficult to inactivate by alcohol or soap.
The most important of these is a protein called spike protein. Virus uses the spike protein to achieve specific adherence to host cells. The inner part of the envelope is a protein shell structure called the capsid. In the case of non-envelope viruses, the outermost layer is the capsid.
So, spike protein is embedded there. Capsid contains genomic nucleic acid, i.e., DNA or RNA.
3. Apearance formed by Capsid protein
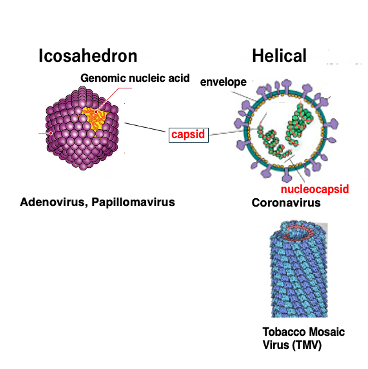
Let’s look at the appearance of the shell formed by the capsid protein. Capsid proteins form icosahedral or helical three-dimensional structures.
The icosahedral capsid is formed by the regular assembly of small proteins. Genomic nucleic acids are stored in the shell. Viruses with such a capsid include Adenovirus and Papillomavirus.
On the other hand, direct attachment of the capsid protein to genomic DNA or RNA forms a helical nucleocapsid, i.e., a complex of nucleic acid and capsid protein. The attachment of the protein to the nucleic acid results in the appearance of a helical capsid. An example of such is the Coronavirus or Tobacco mosaic virus.
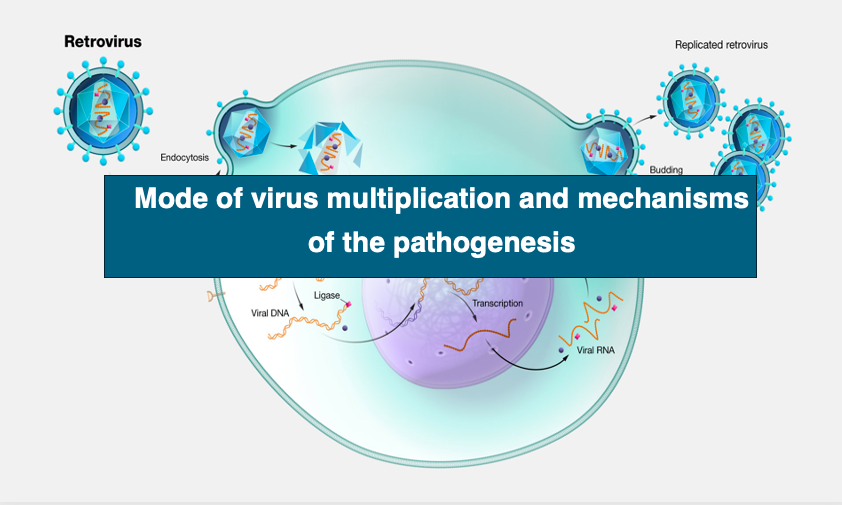
4. Process of initial infection of envelope virus

Now we will look at the process of initial infection of an envelope virus into a host cell.
First, if the host cell is eukaryotic, the virus is specifically adsorbed to the hose cell surface using spikes. Then, virus particle enters the cell by endocytosis. Following it, the capsid is released into the cytoplasm by fusion of the host cell’s lipid bilayer with the envelope’s membrane.
On the other hand, if the viral host is a bacterium, the capsid is released directly into the host cytoplasm by fusion of the envelope with the bacterial cytoplasmic membrane.
5. Process of initial infection of non-envelope virus
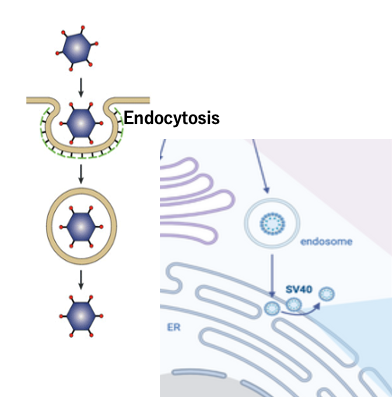
In the case of non-envelope viruses, the capsid is the outermost layer. The capsid attached to the host surface invades the cytoplasm by endocytosis.
In the cytoplasm, the outermost layer is the membrane made of the host’s cell membrane.
There are several ways in which viral particles can be released from this membrane. For example, in the case of SV40, the endocytosis membrane fuses with the ER membrane to release viral particles into the cytoplasm.
Virus Classification
Viruses are classified into seven categories based on the nucleic acid (DNA or RNA) in the virus particle and the genome replication method
6. Names given to DNA and RNA strands
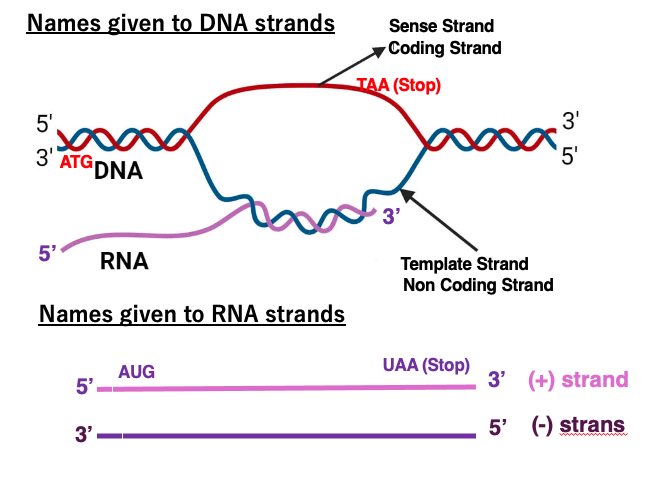
DNA and RNA strands have various names, so let’s review them.
First, this blue DNA strand is called the template strand because it serves as a template for transcription. The base sequence of the messenger RNA generated by transcription of this template strand, and the base sequence of the red strand are very similar. The difference is uracil and thymine. Therefore, the red strand is called coding strand or sense strand. On the other hand, the blue template strand is also called a non-coding strand.
Here are the names given to the RNA strands. This strand is normal messenger RNA. It has AUG initiation and UAA termination codons. The mRNA strand with the amino acid sequence information is called the plus strand. In contrast, an RNA strand that has a complementary sequence to the plus strand is called a minus strand.
7. Four categories of RNA viruses
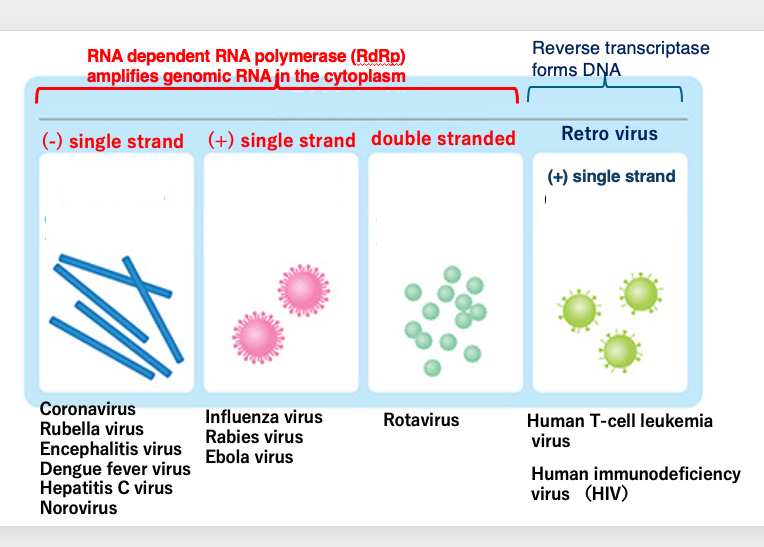
Now we start to study how RNA viruses are classified. Viruses that have RNA as their genome within the viral particle are referred to as RNA viruses, and RNA viruses are classified into four types. Three of them have their genomes amplified in the cytoplasm by RNA dependent RNA polymerase.
The other type is called retro virus, which forms DNA as a replication intermediate by reverse transcriptase, and then forms DNA with long terminal repeats at the both ends. Such viruses have plus single-stranded RNA as the genome.
Viruses whose RNA genome is amplified by RNA-dependent RNA polymerase are classified into three types: minus-single-stranded, plus-single-stranded, and double-stranded RNA types. Examples with minus single-stranded RNA include Coronaviruses and Rubella viruses. Influenza and Rabies viruses are examples of RNA viruses with plus-single-stranded genomes, and Rotaviruses are known to have double-stranded RNA genomes.
Retroviruses that increase by forming DNA intermediates through reverse transcriptase include human T-cell leukemia virus and human immunodeficiency virus.The genome of RNA viruses is linear, regardless of whether it is single-stranded, double-stranded, plus-stranded, or minus-stranded.
8. Three categories of DNA viruses
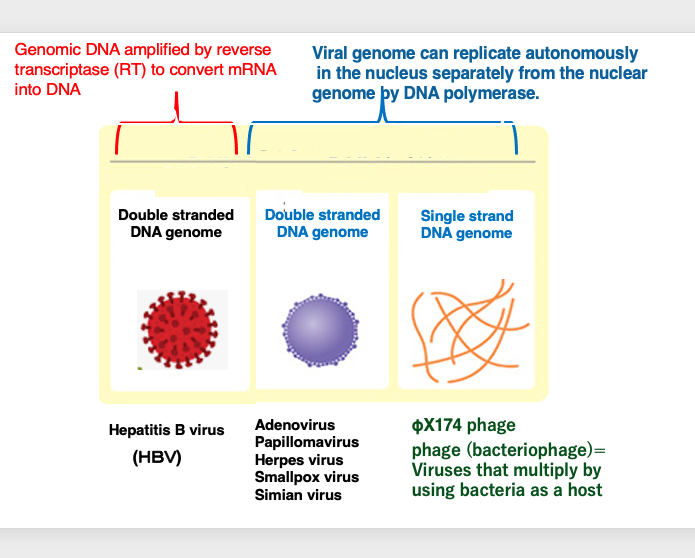
DNA viruses, which carry their genetic material in the form of DNA particles, can be classified into three groups.
Firstly, DNA viruses that independently replicate their own genomes within the host cell nucleus through the action of DNA polymerase can be divided into two groups.
One group consists of viruses that have double-stranded DNA as their genome, such as adenoviruses, smallpox virus, and SP40. The other group comprises viruses that have single-stranded DNA as their genome, such as the ΦX174 phage.
The term “phage” refers to viruses that infect and replicate within bacterial hosts, as opposed to those that infect eukaryotic cells.
The third group of DNA viruses consists of those that have double-stranded DNA as their genome but utilize reverse transcriptase to convert messenger RNA into DNA, thereby facilitating genome replication. The hepatitis B virus is an example of this type of DNA virus. Thus, DNA viruses can be classified into three groups based on their genetic material and replication mechanisms.
9. Classification of DNA viruses by genomic DNA structure
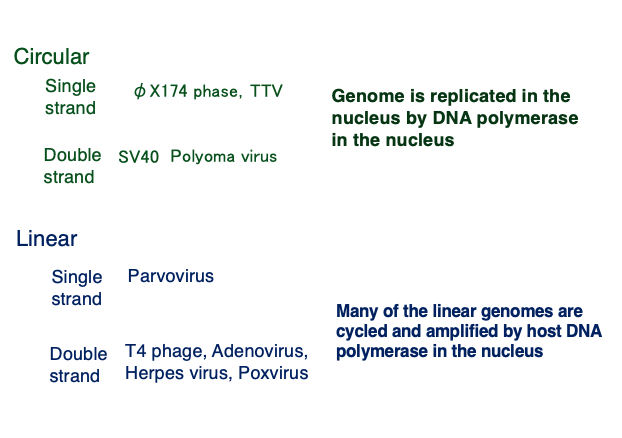
Based on the structure of their genomic DNA, DNA viruses can be classified as shown in this table.
Examples of viruses with a circular single-stranded genome include φX174 phage. Examples with a circular double-stranded genome include SV40 and Polyomaviruses. These circular DNA viruses replicate in the host nucleus using the host’s DNA polymerase starting from the ori sequence.
An example of a virus with a linear single-stranded DNA genome is Parvovirus. Examples of viruses with linear double-stranded DNA genomes include T4 phage, Adenoviruses, and Herpesviruses. Many of these linear genomes circularize and replicate in the nucleus using the host’s DNA polymerase.
10. Carcinogenesis by DNA virus infection
I would like to explain about carcinogenesis caused by various viral infections.
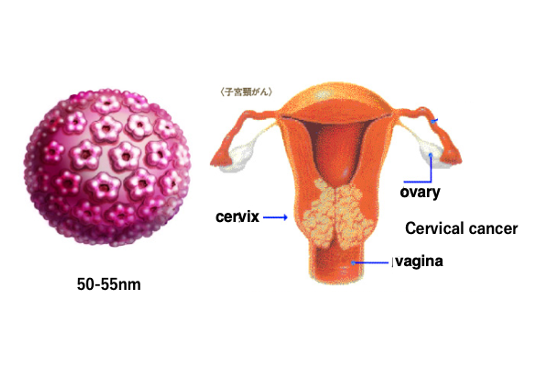
First, I will explain the carcinogenesis caused by DNA viral infections. As an example, I will explain the development of cervical cancer caused by infection with the papilloma DNA virus, which has a circular double-stranded DNA.
Papillomavirus can infect the cervix and cause the development of malignant warts. It is believed that over a long period of time, these warts become cancerous.
It takes a very long time from the infection of the virus to the development of cancer, taking from 10 to 15 years. Cervical cancer is now the second leading cause of cancer mortality in women. The size of the papillomavirus is about 50 – 55 nm in diameter.
11. Genomic structure of HPV
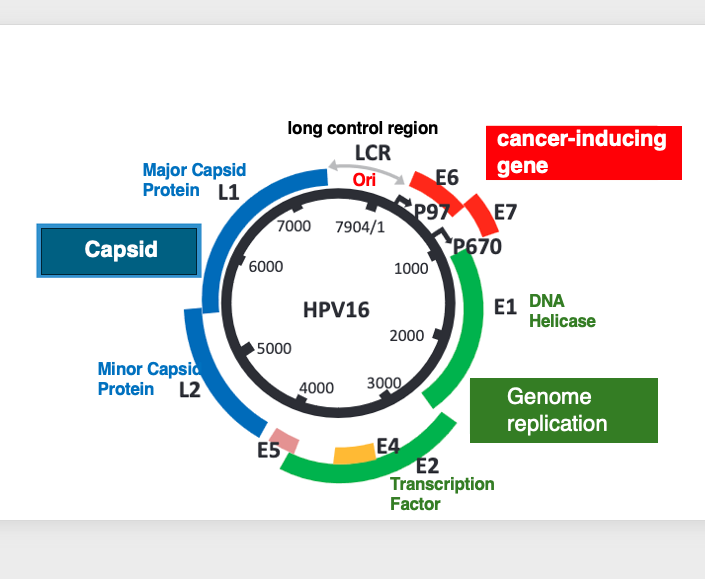
The human Papillomavirus Type 16, which causes malignant cervical cancer, has a circular double-stranded DNA genome.
All the protein-coding regions are located on one strand. The genes encoded in the genome include L1 and L2, which code for the viral capsid proteins. There are also E1 and E2 genes involved in genome replication, with E1 coding for a DNA helicase and E2 coding for a transcription factor.
Additionally, the E6 and E7 genes are known to be involved in causing cancer. The genome also contains an origin of replication (ori) sequence for DNA replication.
12. Carcinogenesis caused by HPV infection
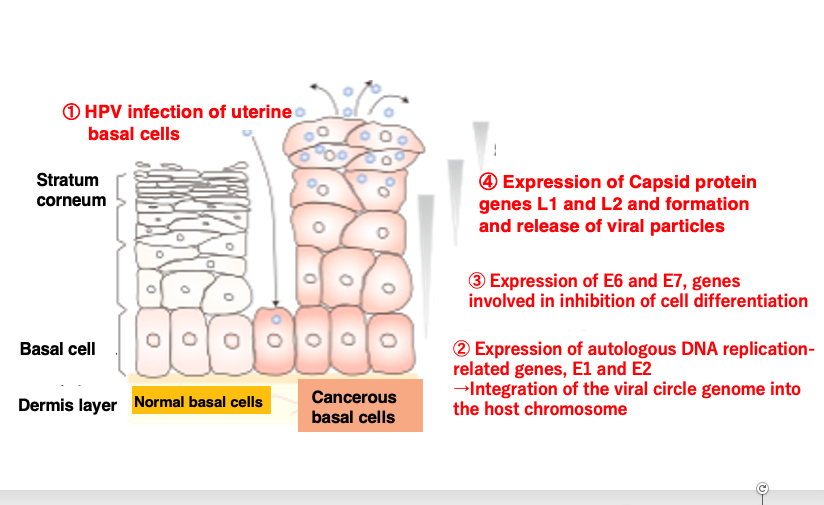
The human papillomavirus, HPV infects the basal cells located beneath the stratum corneum and above the dermis layer through small wounds or abrasions.
Once infection is established, it expresses the E1 and E2 genes related to self-genome DNA replication, increasing the number of its circular genomes and eventually integrating them into the host chromosome.
Subsequently, it expresses the E6 and E7 genes involved in suppressing host cell differentiation and promoting carcinogenesis. Despite these genes expression, as the infected basal cells differentiate over time into the non-dividing stratum corneum, the virus expresses the L1 and L2 capsid protein genes essential for viral particle formation, assembling viral particles within the host cells and releasing them to neighboring cells.
The released virions can then re-infect basal cells through wounds or abrasions, perpetuating this cycle. During this process, carcinogenesis of the basal cells can occur.
13. Creation of a favorable environment for viral proliferation
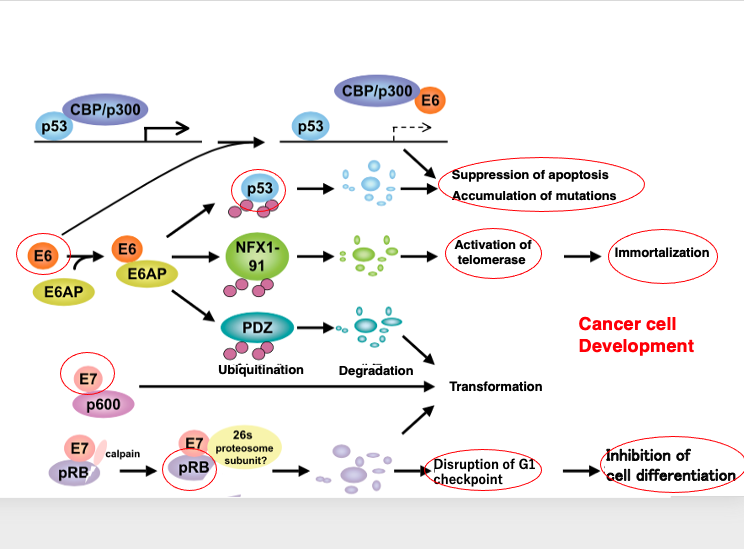
Human papillomaviruses (HPVs) play a crucial role in cellular carcinogenesis. Let’s check the detailed process of it. These viruses possess oncogenes, such as E6 and E7, that contribute to cancer development. The E6 gene encodes a protein that interacts with p53, inhibiting its function and leading to the suppression of apoptosis and the accumulation of mutations. Additionally, E6 activates telomerase, promoting cellular immortalization
The E7 gene encodes a protein that binds to the retinoblastoma (RB) protein, disrupting the G1 checkpoint of the host cell cycle. This results in the inhibition of cellular differentiation and the promotion of immortalization. This results in the inhibition of cellular differentiation and the promotion of immortalization. Through the concerted actions of the E6 and E7 proteins, Human papillomaviruses induce carcinogenesis in the infected cervical cells, creating an environment appropriate to their active replication.
Onset of disease due to RNA virus infection amplified by RNA dependent RNA polymerase
Severe Acute Respiratory Syndrome (SERS) due to coronavirus (covid-19) infection
14. Amplification of viral RNA genome by RNA dependent RNA polymerase
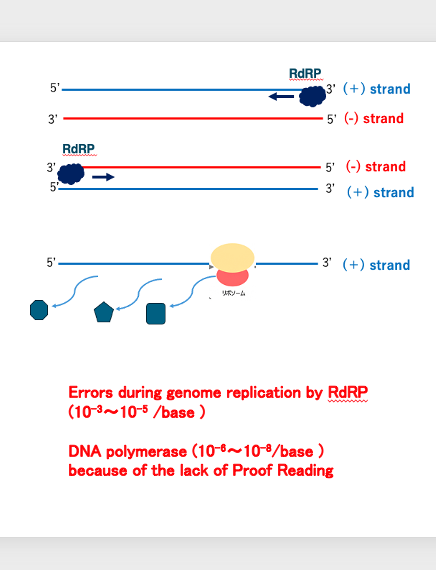
Now, I will explain the amplification of viral genome by RNA dependent RNA polymerase.
For example, in the case of this virus, what is contained in the viral particle is a positive strand of RNA. I have already explained the meaning of the plus strand. Positive strand can be translated by ribosome to produce proteins.
It means that the RNA chain has information for this purpose. When a new strand is synthesized by RNA dependent RNA polymerase using the positive strand as a template, the resulting strand is a negative strand with a complementary base sequence to the positive strand.
On the other hand, in the case of this virus, if the particle contains a minus strand, the RNA strand produced by RNA dependent RNA polymerase is a plus strand. The error ratio in replication by RNA dependent RNA polymerase is said to be 10-3〜10-5 /base.
This is approximately 1000 times higher than the replication by DNA polymerase, which is 10-6〜10-8/base. This high error rate for replication by RNA dependent RNA polymerase is due to the absence of a Proof Reading mechanism.
15. Structure of coronavirus
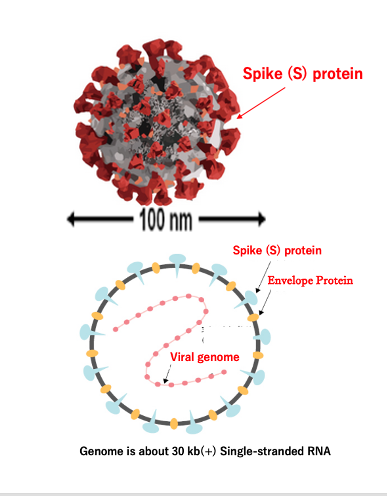
Let me explain the basic structure of the coronavirus. This is the appearance of the coronavirus, with spike proteins embedded in the envelope that are involved in specific binding to the host cell. The diameter is around 100 nanometers.
Here I show the internal structure. The spike proteins and E proteins are embedded in the envelope. The capsid proteins directly bind to the positive-sense single-stranded RNA genome. The size of the genome is around 30 kilobases.
This is the basic structure of the coronavirus. It is a virus with an exceptionally large RNA genome, which is replicated by an RNA-dependent RNA polymerase. However, this RNA virus has an unusual proofreading mechanism. Therefore, despite its large genome size, it can replicate with a certain degree of accuracy.
16. Detailed of Coronavirus structure
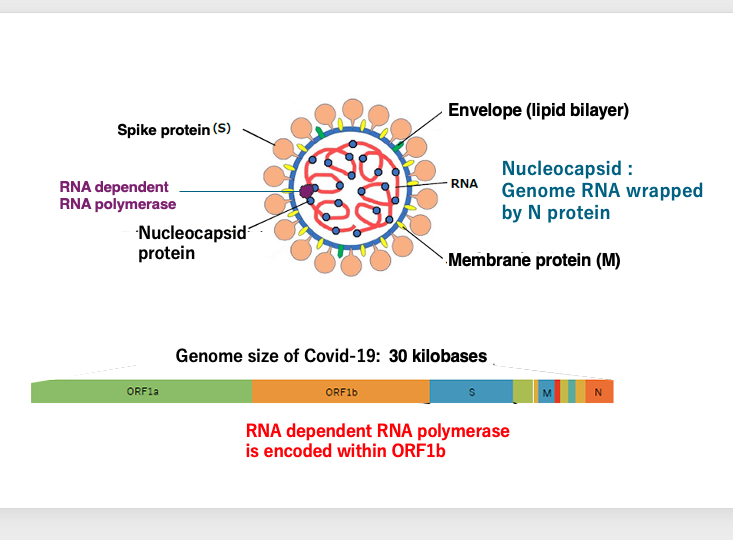
Here is a more detailed description of the coronavirus structure. In addition to the S protein, the envelope contains embedded N and M proteins.
The viral particle contains pre-packaged RNA-dependent RNA polymerase proteins.
This is the genome structure of Covid-19, a recently prevalent type of coronavirus. The ORF1B region encodes the RNA-dependent RNA polymerase. Additionally, the S protein, M protein, N protein, and others are coded on this genome.
17. Symptoms of Coronavirus infection compared to common cold
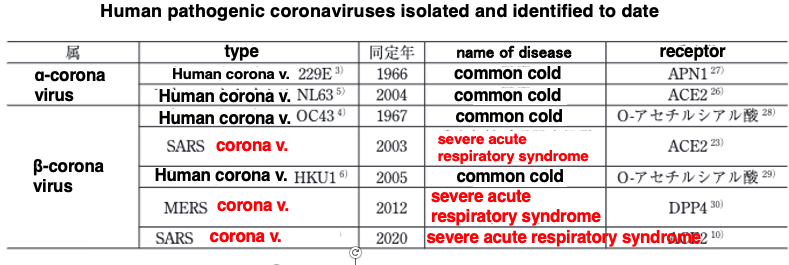
Here is a summary of the symptoms of coronavirus infection compared to the common cold: We usually catch what is called with mild symptoms like runny nose, sore throat, and cough. It is said that 10% to 10% of such colds are caused by four types of coronaviruses that infect people on a daily basis. When infected with a human coronavirus such as 229E or OC43, NL63, or HKU1, which are routinely transmitted among human, they can cause symptoms of the so-called common cold.
On the other hand, in 2003, a coronavirus that seemed to use bats as its natural host infected humans. When a person is infected with a coronavirus whose natural host is not a human, he or she can develop a very serious respiratory illness.
In 2012, a coronavirus, whose natural host was the Camel, infected humans and was named Middle East Respiratory Syndrome, or MERS, because it spread primarily in the Middle East. This was also accompanied by very severe respiratory illnesses.
The most recent outbreak was in 2020. A coronavirus, probably with bats as the natural host, infected humans, causing severe respiratory illness. Thus, when a coronavirus crosses the species barrier and infects humans, it causes a very serious respiratory disease.
18. Coronavirus Infection to cells
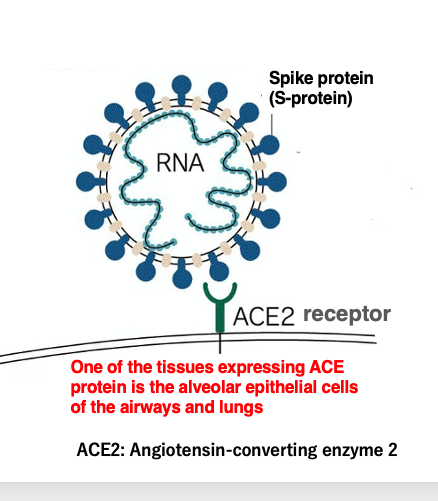
We will discuss the first reaction of coronavirus infection: adsorption to human cells in detail. The spike protein embedded in the coronavirus envelope can bind specifically to the ACE receptor protein, which is a metalloproteinase receptor.
This receptor is expressed in large numbers in the alveolar epithelial cells of the airways and lungs. So, coronaviruses will infect alveolar epithelial cells in the lungs. Amino acid mutations in the spike protein have a major impact on the infectivity of coronaviruses.
For this reason, coronavirus mutations are often described with a focus on amino acid mutations in the spike protein. For example, when a strain is described as E484K mutant, it means that the 484th amino acid of the spike protein, glutamic acid, E, has been mutated to lysine, K.
19. Proliferation cycle of Corona virus
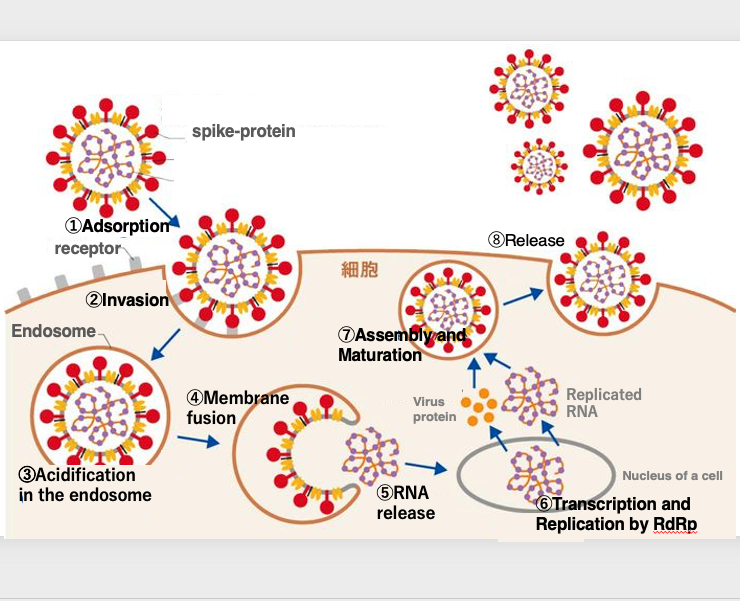
Next, I will explain the replication cycle of the coronavirus. After the coronavirus attaches to the alveolar epithelial cells, it is engulfed into the cytoplasm by endocytosis.
As the pH inside the endosome becomes acidic, the outer endosomal membrane fuses with the inner envelope membrane, releasing the genomic RNA into the cytoplasm.
The genomic RNA enters the nucleus, where its replication is carried out by the RNA-dependent RNA polymerase.
The RNA is then translated in the cytoplasm to produce the proteins necessary for constructing the virus particles.
As the viral proteins assemble and the envelope is formed, the genomic RNA is incorporated inside, and eventually released from the cell surface membrane. This is the replication cycle of the coronavirus.
20. Therapeutic agent: Inhibitor of RNA dependent RNA synthetase
Remdesivir is one of the treatments for coronavirus infection. Remdesivir was originally developed as a treatment for Ebola virus.
The Ebola virus has a negative single-stranded RNA genome. Remdesivir was developed as a drug that inhibits the strand elongation reaction by the RNA-dependent RNA polymerase in this virus.
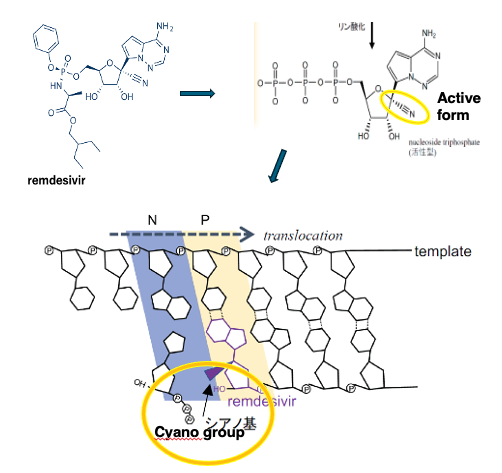
Here is the structural formula of remdesivir. In the cell, remdesivir is metabolized into a nucleotide triphosphate analog with a characteristic cyano group.
When remdesivir is incorporated by the RNA-dependent RNA polymerase, the steric hindrance caused by the cyano group prevents the polymerase from incorporating and binding the next nucleotide triphosphate. Through this mechanism, remdesivir inhibits the replication of the coronavirus.
21. Infection and Development of Retroviruses with a long latency period
Let me explain the infection and pathogenesis of retroviruses, which have a long incubation period. First of all, retroviruses have single-stranded plus-stranded RNA as a game.
However, the genome is amplified not by RNA dependent RNA polymerase but by reverse transcriptase. DNA produced by reverse transcription has a long terminal repeat, LTR. The DNA with this long terminal repeat has the step of being embedded in the host genome.
One such retrovirus is the AIDS virus, Human Immunodeficiency Virus, or HIV virus, which has a long asymptomatic period of 5 to 10 years, a latent period, and causes a decrease in immunity due to a low white blood cell count.
As acquired immunodeficiency syndrome develops, this can lead to increased susceptibility to pneumonia, malignancy, and death.
22. Structure of HIV Particle
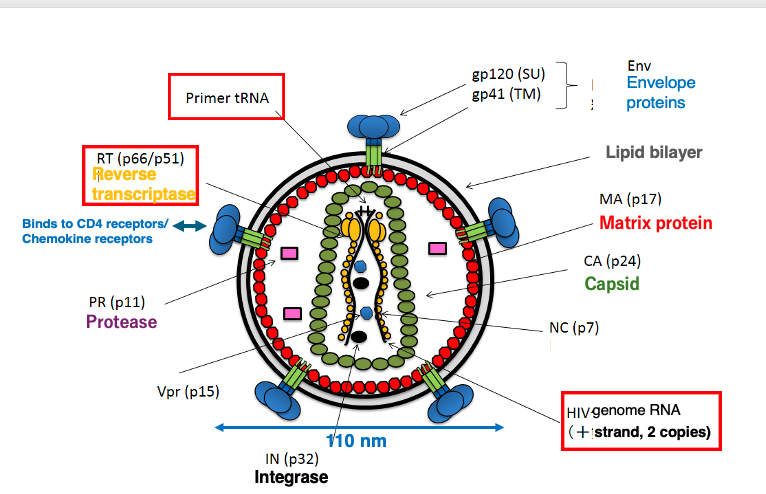
I will explain the basic structure of the HIV virus. It has an envelope with embedded spike proteins. These spike proteins specifically bind to the CD4 receptor and chemokine receptors on the host cell surface.
Regarding the genome, it contains two copies of a single-stranded positive-sense RNA genome. The viral particle also contains a reverse transcriptase enzyme to transcribe the RNA genome into DNA. Additionally, the particle incorporates a tRNA molecule that serves as a primer for the reverse transcription reaction.
The green color represents are the capsid proteins. The RNA genome, reverse transcriptase enzyme, and tRNA primer are encapsulated within the capsid shell.
This is the basic structure of the HIV virus. Its size is approximately 110 nm in diameter.
23. HIV genome ognamization (~ 9.5 Kb)
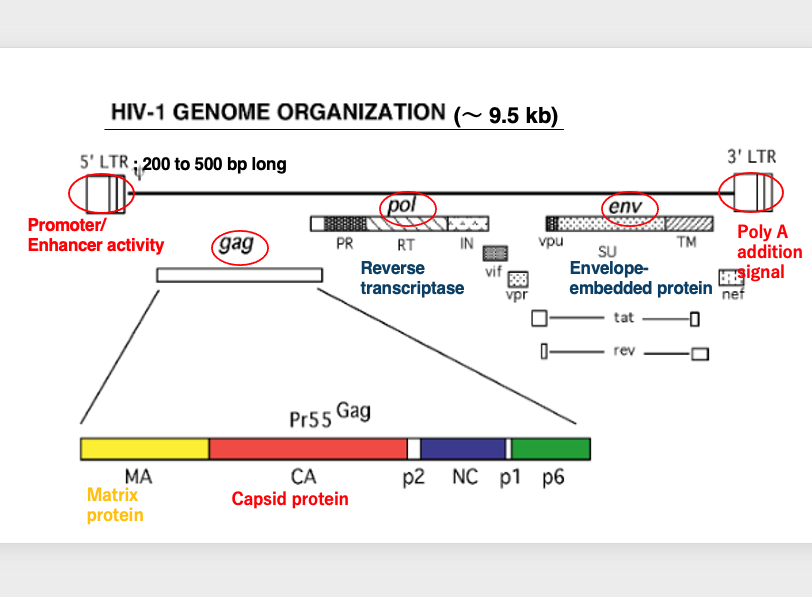
Now, I will explain the structure of the HIV viral genome. This shows the structure of the reverse transcribed HIV genome.
First, there are long terminal repeats. The 5′ long terminal repeat has promoter activity and enhancer activity. On the other hand, the 3′ long terminal repeat contains a polyadenylation signal.
There is also a large protein coding region called GAG. The GAG region includes matrix proteins and capsid proteins. Next, there is the POL region. This region encodes the reverse transcriptase enzyme and others. Finally, there is the ENV region. This region encodes the envelope protein and others.
Overall, the HIV virus has a length of approximately 9.5 kilobases in its reverse transcribed DNA form.
24. Cells that HIV infects
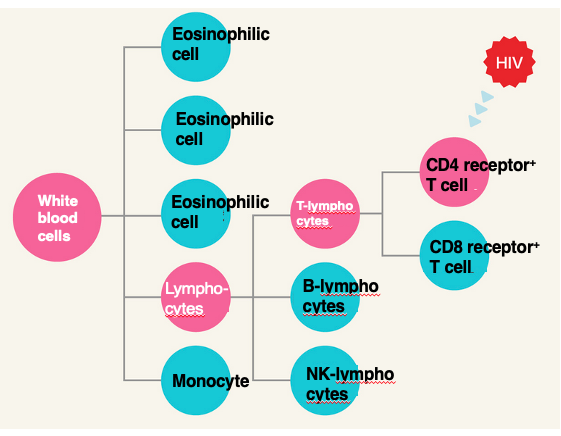
Here, this shows the cell groups that raised from white blood cells. Various cells can be formed by the differentiation of white blood cells.
Lymphocytes are one of them. Lymphocytes further differentiate into T lymphocytes and B lymphocytes. Additionally, one of the cells that can be formed by the differentiation of T lymphocytes is the CD4 receptor positive T cell, which expresses the CD4 receptor.
The HIV virus infects this CD4 receptor positive T cell. This cell plays a central role in the immune system.
25. HIV-Type1 proliferation cycle
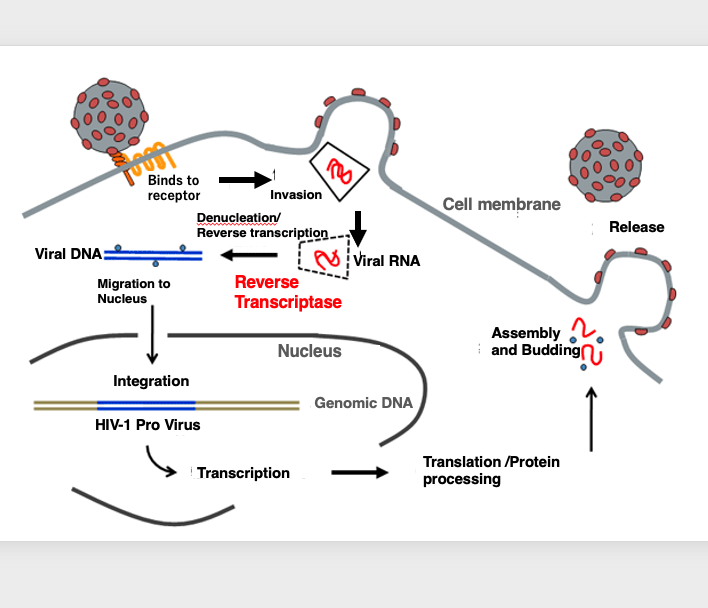
HIV viral replication cycle is described below. The virus first adsorbs specifically to the CD4 receptor. After adsorption is complete, the envelope membrane of the virus fuses with the cytoplasmic membrane of the host, this leads to the release of the capsid shell from the virus. The capsid shell protein is eventually degraded and the viral RNA genome contained within is released into the cytoplasm.
The RNA genome is converted to double-stranded DNA by reverse transcriptase, introduced by the virus itself, and the double-stranded DNA is transferred into the nucleus, where it is subsequently incorporated into the genome. This state of viral integration into the host genome is called, provirus.
In the case of the AIDS virus, the provirus remains in this state for a long period of time, from 5 to 20 years. During this time, there is no active transcription of viral genes. Therefore, a latent period of disease is followed by a less pronounced disease state.
Viral transcription becomes active when the host’s physical condition changes, for example. As the viral RNA is translated, the proteins necessary to reconstitute viral particles are stored in the cytoplasm, from which many viral particles are formed. They are eventually released from the cell surface. This is the HIV viral replication cycle.
26. Onset of AIDS : Changes in the number of CD4-positive lymphocytes
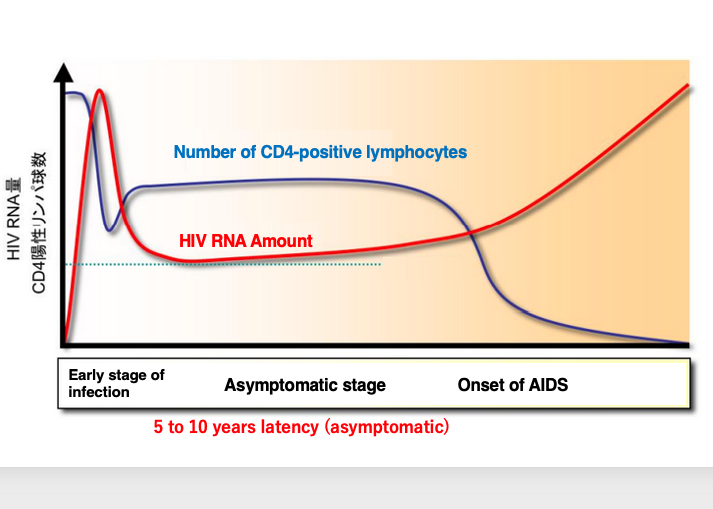
This figure summarizes the changes in the number of CD4+ lymphocytes from HIV infection to the onset of AIDS. Immediately after HIV infection, the number of CD4+ lymphocytes initially decrease, but is then maintained at a certain level.
This period lasts from 5 to 10 years, and in some cases up to 20 years. During this period, no obvious symptoms appear, so it is called the asymptomatic period. Subsequently, triggered by events such as a significant change in the host’s condition, HIV transcription becomes more active.
As the amount of HIV increases, the number of CD4+ lymphocytes, which are the infected cells, decreases rapidly. The decrease in these cells, which play a central role in the immune response, leads to the onset of various conditions caused by immunodeficiency.
27. Onset of AIDS due to decrease in CD4-positive cells
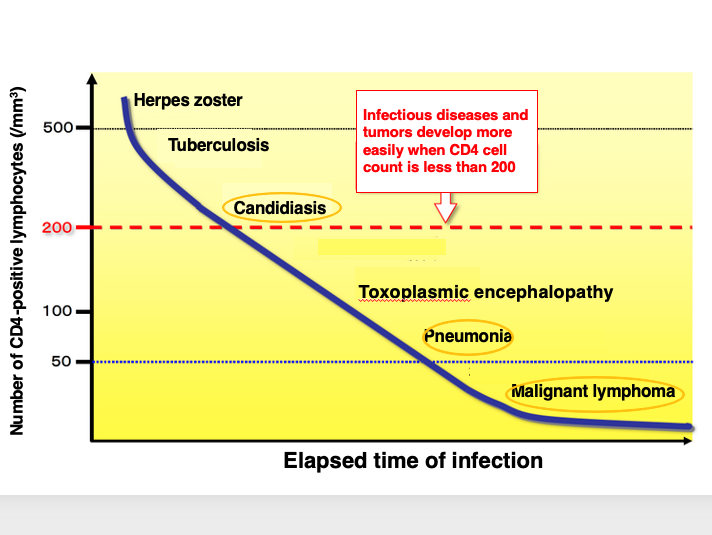
In this figure, the disease occurs as a result of a decrease in CD4-positive cells are summarized.
When the number of CD4-positive cells drops to about 500 cells per milliliter, shingles and other symptoms appear, and the patient is more susceptible to tuberculosis and other infections. When the number of CD4-positive cells drops to around 200, the patient becomes susceptible to candidiasis, an opportunistic infection.
If the level falls below 50, the patient is more susceptible to pneumonia, etc. If the level falls well below 50, malignant lymphoma may develop. In this way, the HIV virus infects and destroys CD4-positive cells, which are at the center of the immune response, thereby escaping the immune response and maintaining an environment, in which it can easily multiply within the host’s cells.
28. Retroviruses with cell-derived oncogenes into the genomes: Rous Sarcoma Virus
Let’s change the topic a bit, and I will explain retroviruses that carry host-derived oncogenes. In certain cancer tissues, specific retroviruses can be found. Moreover, those retroviruses may have host-derived oncogenes embedded in their genomes. As an example of this, I will discuss the Rous Sarcoma Virus.
29. Reformation of Sarcoma by inoculation of crushed sarcoma cells filtrate

In 1911, Rous succeeded in reforming sarcoma by ingesting a filtered extract of chicken sarcoma tissue into normal individuals. First, he crushed chicken sarcoma tissue and filtered it through a ceramic filter. The filtrate contained a virus. By ingesting the filtrate into chickens, new sarcomas were formed in this experiment.
30. Genome structure of Retro virus
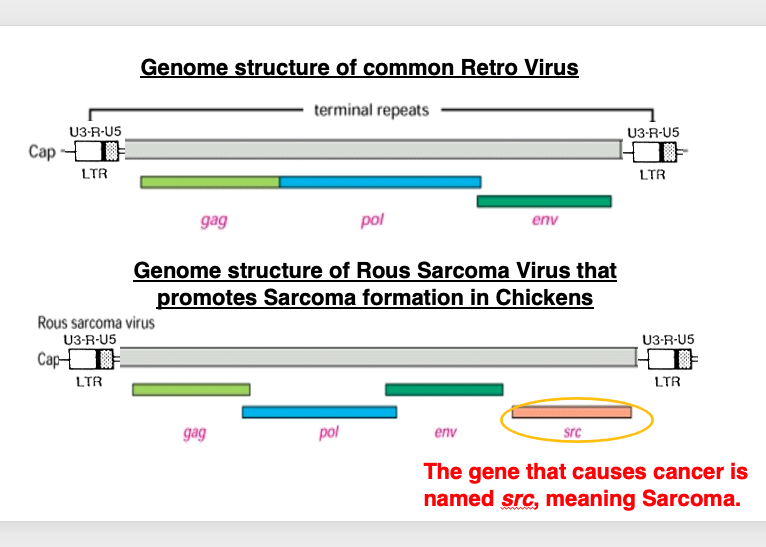
This slide compares the genome structure of a common retrovirus, with that of Rous sarcoma virus, which causes sarcoma in chickens. The genome structure of a common retrovirus has long terminal repeats at the both ends. There are three large regions that encode genes: Gag, Pol, and Env. On the other hand, in the case of Rous sarcoma virus, there are long terminal repeats at both ends and three protein coding regions, Gag, Pol, and Env, as in the above case, but a src gene is found downstream of the Env region.
This src gene is named so, meaning a gene that has potential to cause sarcoma.
31. Viral src gene (V-src) homologous one (C-src) in the host genome
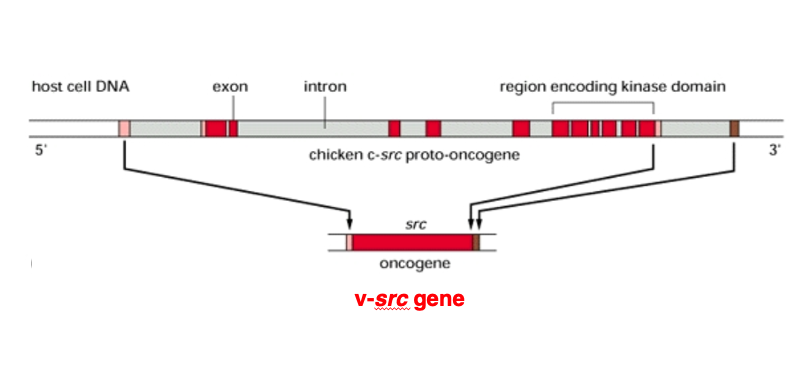
After subsequent research, a gene homologous to the SRC gene found in the Rous sarcoma virus genome was found in the host cell genome.
The cellular counterpart of the SRC gene, called c-SRC, has an exon-intron structure. In contrast, the viral SRC gene, termed v-SRC, lacks introns as it represents the spliced form of the gene after intron removal.
Currently, to distinguish these two forms, the viral gene is referred to as v-SRC, while the cellular gene is called c-SRC.
32. Oncogenes found within tumor-causing retroviral genomes
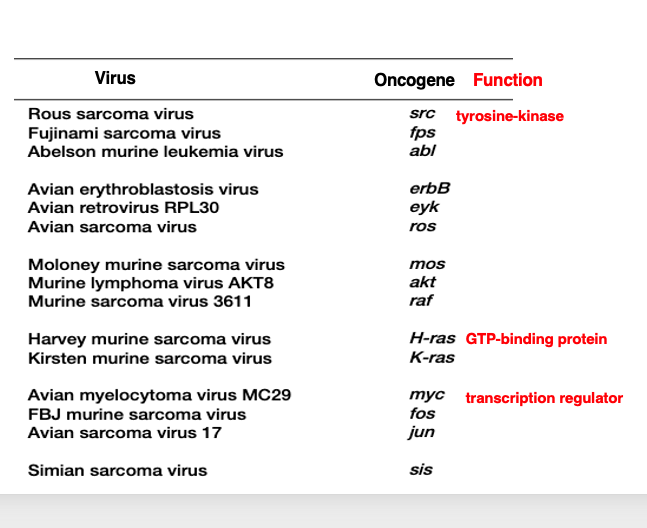
Subsequently, similar studies continued, and many oncogenes were discovered in retroviruses. This is the Rous sarcoma virus I mentioned earlier. It contained a gene called src, which is now known to encode a tyrosine kinase.
In the case of Harvey murine sarcoma virus, it has the ras oncogene, which is known to be a GTP-binding protein. The Avian myelocytoma virus contains the myc gene, which is a transcription factor.
Thus, for retroviruses that have the ability to cause cancer in the host, it gradually became clear that, their genomes contain host-derived genes involved in cell proliferation and growth control incorporated into the viral genome.
33. Process of retroviruses with oncogenes in their genomes
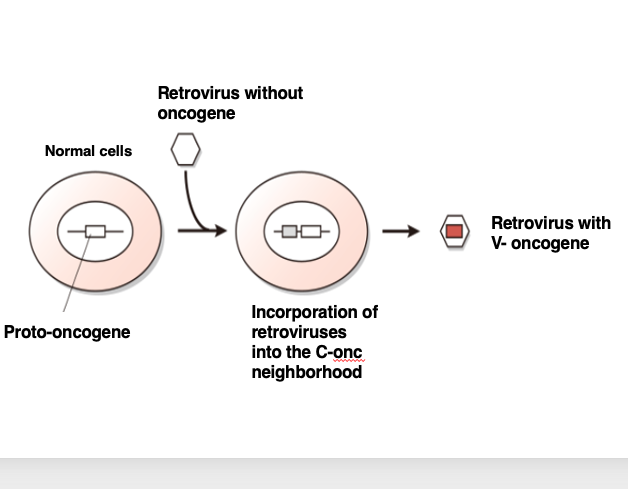
Let me explain the process of how a retrovirus acquires a host oncogene. Here is a normal cell. This cell has genes necessary for its own cell proliferation. When such cell proliferation-related genes are incorporated into a virus, they act as oncogenes. Therefore, genes that act as oncogenes within a virus are called proto-oncogenes, when they are present in a normal cell.
Now, a retrovirus without an oncogene infects this normal cell. Its RNA genome is reverse transcribed into DNA, and integrated into the host genome, and we can assume that the viral DNA happened to be integrated near a proto-oncogene.
Subsequently, the virus transcribes its own DNA to produce an RNA genome, forms viral particles, and exits the cell. In this process, the viral genome incorporates the oncogene as part of its own genome, leading to the establishment of a retrovirus with an oncogene.
By acquiring an oncogene in its genome, the retrovirus gains the ability to transform the host cell into a cancer cell, thereby now the virus can create an environment that allows efficient long-term replication of the viral genome.
34. Process of Rous sarcoma virus with src gene
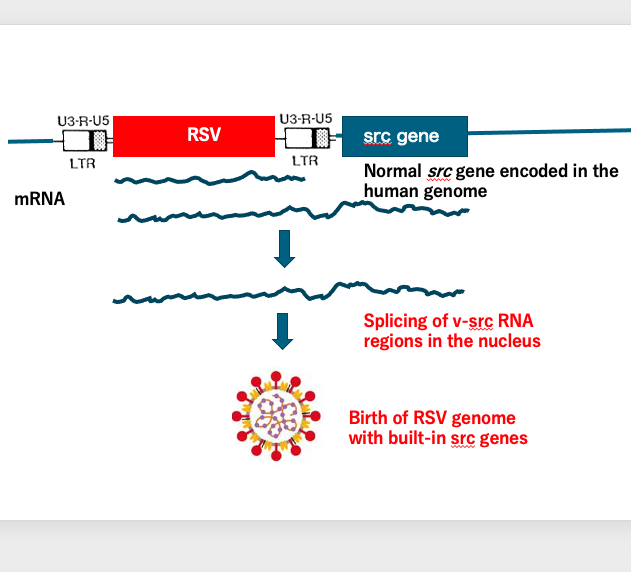
The birth of Rous sarcoma virus carrying the intronless src gene can be explained as follows: By chance, the Rous sarcoma virus genome was integrated upstream of the host’s src gene.
Normally, transcription of the Rous sarcoma virus starts from the upstream long terminal repeat (LTR) and terminates at the downstream LTR. However, in this case, transcription continued beyond the downstream LTR and into the src gene, terminating downstream of it. This resulted in an unusually long transcript. This transcript underwent splicing in the nucleus.
While the Rous sarcoma virus genes originally lacked introns, the host’s src gene contained many introns. Through splicing, Rous sarcoma virus with an intronless src gene was produced.
This intronless src-containing RNA was then packaged into the viral particle, giving rise to the Rous sarcoma virus that we observe today, carrying the oncogenic src gene.
Well, I will end this lecture now. See you again soon.
コメント