This lecture can be viewed as a video on YouTube site: Life Scinence Lectures for you
1. Contents of this lecture
This lecture explains the rRNA and ribosomal proteins that make up the ribosome. This lecture contains 17 figures.
- It describes the rRNAs contains in the large subunit and the small subunit, as well as the positional relationships of the proteins located there. It also mentions the operons that code for rRNA genes and the operons coding for ribosomal proteins.
2. This lecture provides a detailed explanation of the movements of the large and small subunits of the ribosome during the elongation reaction of the peptide chain, and the positions of peptidyl-tRNA and aminoacyl-tRNA within the ribosome.
3. Additionally, it refers to the fact that rRNA is a ribozyme with catalytic activity for the peptide transfer reaction.
Key words: E-site,P-site, A-site, Large subunit, Small subunit, 5S rRNA,1 6S rRNA, 23S rRNA, Ribozyme, Peptidyl-tRNA, Aminoacyl-tRNA, Large Subunit, Small subunit, Aminoacyl-tRNA, Peptide transfer, Nucleolus, Ribosome assembly, L7/L12,35S rRNA, Ribosomal RNA operon, tRNA, hybrid site
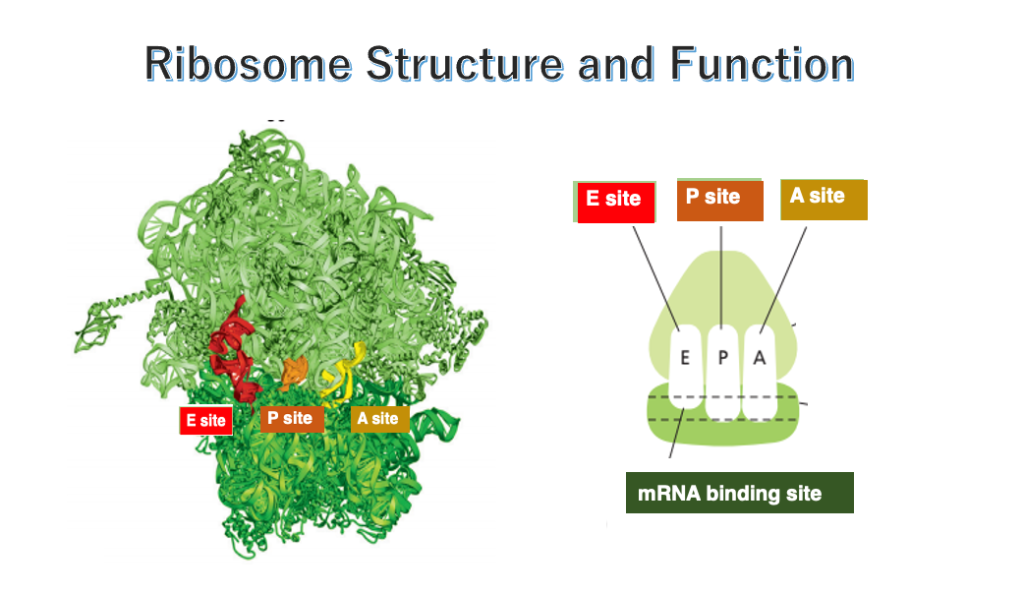
2. Ribosome: produces peptide according to messenger RNA information
Ribosome is a device that produces peptide chain according to information of messenger RNA, by connecting amino acids together in the order specified by codons.
In order to carry out this reaction smoothly, the ribosome has three cavities that each of them can hold a transfer RNA, which are called the E-site, P-site, and A-site. Specifically, transfer RNAs to which amino acids are attached, forms a stable hydrogen bond with an appropriate anticodon, and peptide chain is elongated in a ribosome.
The ribosome has two parts: a large subunit and a small subunit. The large and small subunits form a complex, by placing messenger RNA at the interface between the two subunits.
3. Componet of ribosome
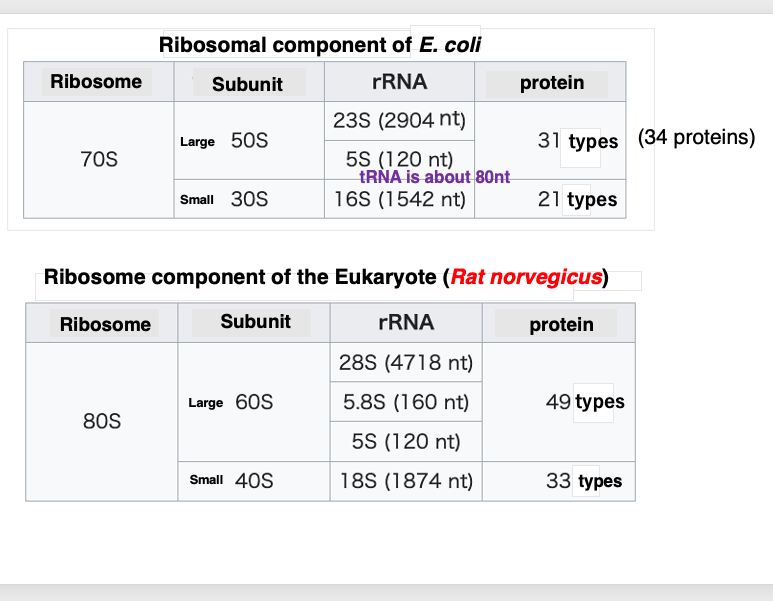
First, I will describe how the ribosome of the bacterium Escherichia coli is structured. The large subunit has a sedimentation coefficient of 50S, and the small subunit has a sedimentation coefficient of 30S. The larger the sedimentation coefficient, the larger the particle and the faster the sediments. The 50S subunit contains two types of RNA. 23S and 5S. 23S RNA consists of 2,904 bases, while 5S has 120 bases. You all know transfer RNA. It consists of about 80 bases.
50S large subunit also contains 31 types of proteins with a total of 34 protein molecules.
On the other hand, 30S small subunit is a complex of a 1,542 base 16S RNA and 21 types of proteins with 21 protein molecules. When the 50S and 30S particles associate, they form a 70S ribosome.
Next, let’s look at ribosomes found in eukaryotic cells. This table shows data for rat ribosomes. Like in prokaryotes, they consist of a large and small subunit. The large subunit is 60S, and the small subunit is a 40S, making them slightly larger than their prokaryotic counterparts.
Eukaryotic large subunit contains three types of ribosomal RNAs , 28S, 5.8S, and 5S. The large subunit also contains 49 types of proteins with 49 protein molecules.
Eukaryotic 40S small subunit contains only one type of RNA called 18S rRNA, which is 1,874 bases long. Additionally, there are 33 types of proteins with 33 protein molecules in this small subunit. When these two subunits associate, they form an 80S ribosome particle, which is slightly larger than the 70S prokaryotic particle.
4. Appearance of ribosomes
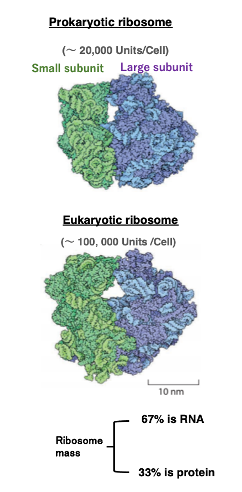
Let’s take a look at the appearance of ribosomes.
On the left is the appearance of a bacterial ribosome. Bacteria have around 20,000 of these ribosomes per cell. On the right is the appearance of a eukaryotic ribosome. Eukaryotes have around 100,000 ribosomes per cell. The green part is the small subunit, and the blue part is the large subunit.
Compared to eukaryotic ribosomes, bacterial ribosomes have fewer constituent proteins, shorter ribosomal RNAs, and therefore, they are smaller in overall size.
While ribosomes are composed of proteins and RNA, in terms of mass, 67% is RNA and 33 % is protein.
5. Two-dimensional model of 16S ribosomal RNA in E. coli
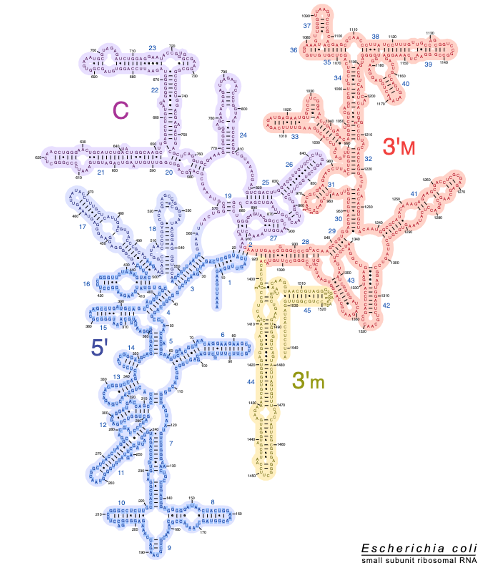
This figure shows a two-dimensional model of 16S ribosomal RNA in E. coli. In reality, ribosomal RNA has a very complex three-dimensional structure. In the two-dimensional structure, these parts, which are represented as hairpins, form hydrogen bonds with other hairpins to form a complex folded three-dimensional structure.
6. Two-dimensional models of 23S and 5S ribosomal RNAs in E. coli
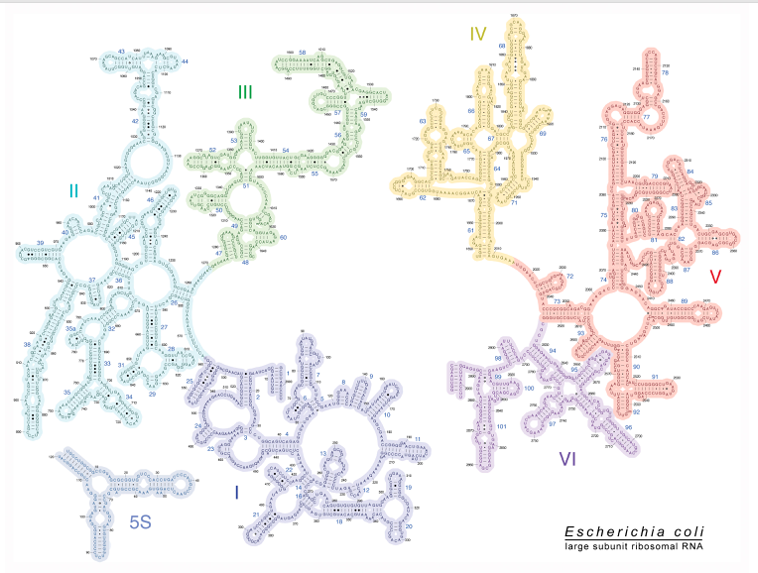
Next, this is a two-dimensional structural model of the 23S ribosomal RNA and 5S ribosomal RNA of E. coli.
In this figure, this part and this part are disconnected, but in reality, they are connected, to form a single RNA molecule. In fact, 23S ribosomal RNA also has a very complicated three-dimensional structure, as it is so for 16S ribosomal RNA.
Here is a two-dimensional model of 5S ribosomal RNA, a short 120-base ribosomal RNA.
7. Three-dimensional look at the structure of a bacterial ribosome
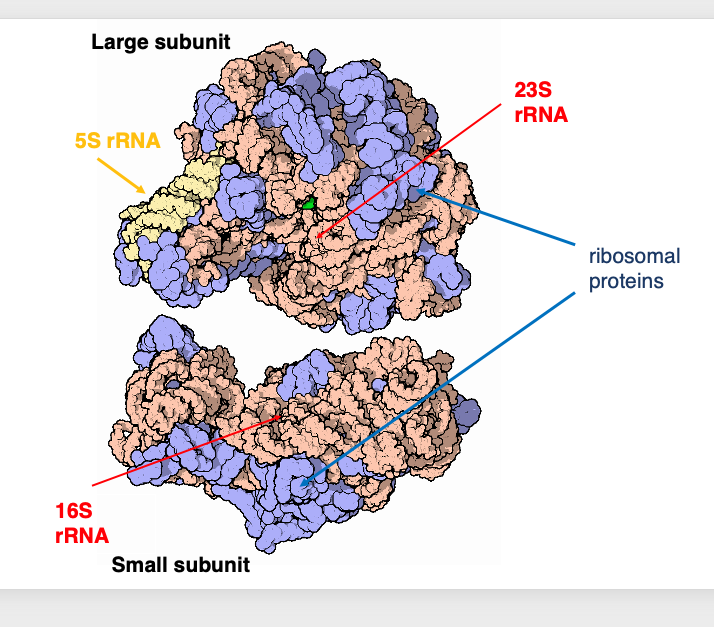
Let’s take a three-dimensional look at the structure of a bacterial ribosome.
This is the large subunit, while this is the small subunit. Here, the 5S ribosomal RNA is shown in yellow. The 23S and 16S ribosomal RNAs are shown in orange. The ribosomal proteins are shown in blue. It is known that ribosomal proteins are not located inside the ribosomal RNA, but rather on the surface of the ribosomal RNA.
The role of ribosomal proteins is to assist in the formation and maintenance of the correct three-dimensional structure of ribosomal RNAs, and to aid in structural changes required for the activation of ribosomal RNAs.
Interestingly, active centers of the various ribosomal reactions are composed of ribosomal RNAs, while proteins do not contain the active centers.
8. Modified ribosomal RNA bases in E. coli
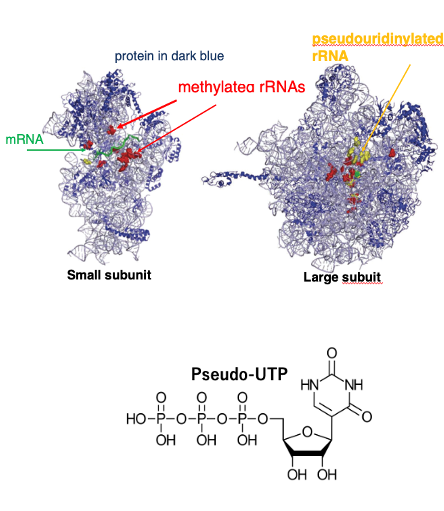
These figures show three-dimensional image of the ribosome of E. coli. This one shows the small subunit, and this one shows the large subunit. In these figures, ribosomal RNA is thinly drawn like this. On the other hand, ribosomal proteins are depicted in dark blue. In ribosomal RNAs.
In these figures, the positions of methylated ribosomal RNA bases are shown in red. In addition, some bases are pseudouridinated. This is Pseudo-UTP and this is UTP.
Such pseudouridinated bases are shown in yellow. It is known that not only RNA bases but also ribosomal proteins undergo post-translational modifications, such as phosphorylation and methylation. These series of modifications are essential for the proper functioning of the ribosome.
9. Architecture of the 80S ribosome of Saccharomyces cerevisiae
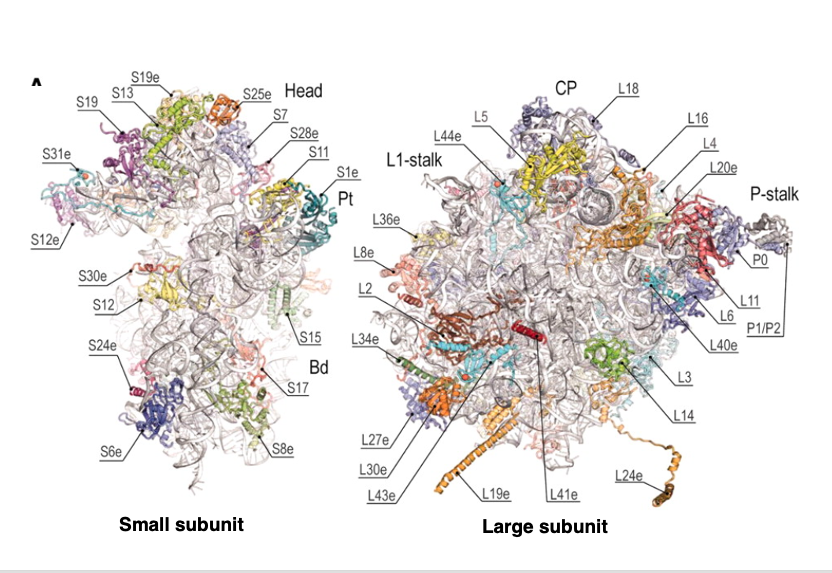
This figure shows the three-dimensional structure of the 80S ribosome of Saccharomyces cerevisiae, a type of eukaryotic yeast. This side shows the small subunit and this side shows the large subunit.
In this figure, the position of the proteins on the surface of the ribosomal RNA is clearly indicated. You can clearly see that the proteins are located on the surface of the RNA.
10. RNA operon structure and processing of the transcript in E. coli
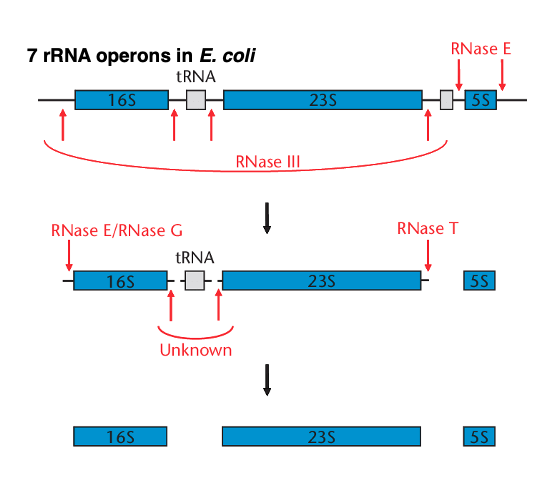
From here on, I will explain how ribosomal RNA and ribosomal proteins are produced. I will explain the transcription of the E. coli ribosomal RNA operon. In E. coli, there are seven ribosomal RNA operons, all of which have similar structures.
Long RNAs transcribed from the operon are fragmented by RNase E, RNase G, RNase T, etc. Finally, this long transcript is processed into 16S ribosome RNA, 23S ribosome RNA, 5S ribosome RNA, and transfer RNA.
11. Negative feedback regulation of ribosomal protein operons
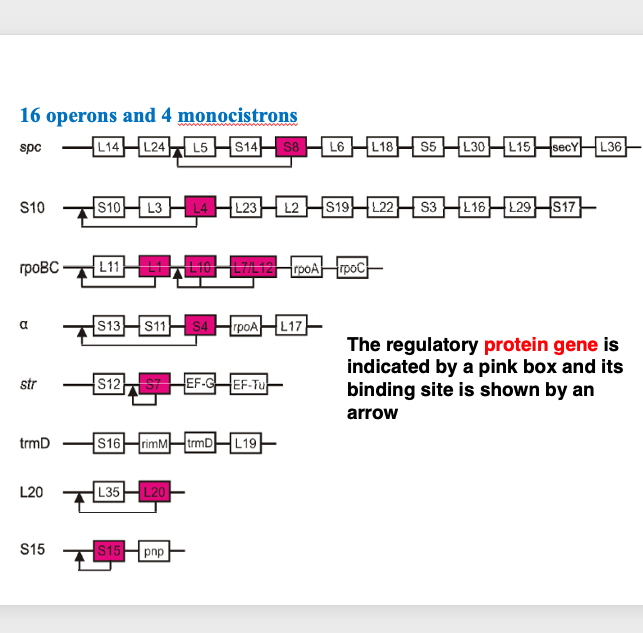
Next, I will explain the operon that codes for ribosomal proteins in E. coli. Regarding the naming of ribosomal proteins, they are generally numbered in order of decreasing molecular weight, with the largest in the large subunit designated as L1, L2, L3, and so on, while in the small subunit, the largest is S1, the next is S2, and so forth.
The ribosomal proteins are encoded by 16 operons and 4 monocistronic transcripts. Here, 8 representative operons are shown. The S8 ribosomal protein gene, shown in red in the spc-operon, is known to function as a regulatory protein by binding to this region of the messenger RNA, exerting negative feedback control.
In the ribosome, each protein is contained one molecule per particle. Therefore, each protein must be produced in precisely the same number. For this reason, precise adjustments are made mainly at the transcriptional level.
Here is a gene described as L7 / L12 gene. In the early days of ribosome research, individual ribosomal proteins were separated by electrophoresis based on their molecular weight differences. At that time, determining the amino acid sequences of peptides was extremely difficult. Now, it is known that the product of theL7/L12 gene exist in the cell in two forms: one with the N-terminus acetylated and one without modification. When separating ribosomal proteins individually by electrophoresis, the modified and unmodified forms appear as proteins with different molecular weights. This is the reason why this gene is named L7 /L12. As mentioned earlier, each protein generally present in one copy per ribosomal, however, it is known that there are 4 copies of L7 and L12 proteins in one large subunit. In the large subunit, L7 and L12 proteins form dimers, and 2 dimers present, so accounting for the 4 molecules.
12. rRNA genes cluster in a yest Saccharomyces cerevisiae
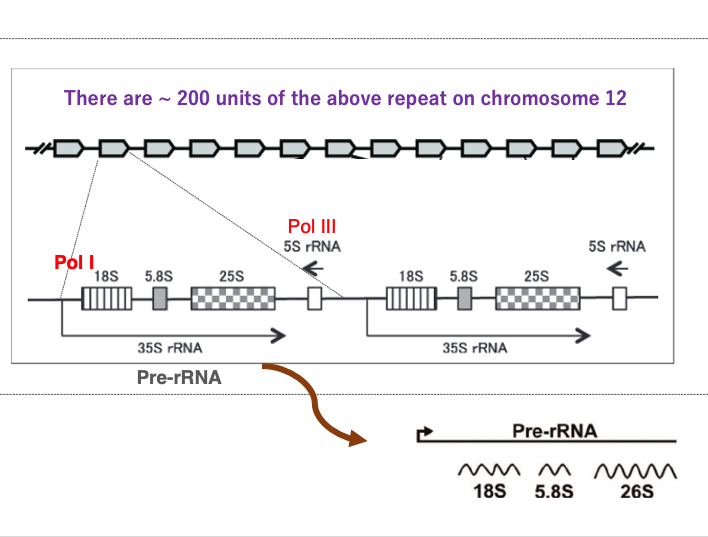
I will explain the transcription of ribosomal RNA genes in eukaryotes, using the yeast Saccharomyces cerevisiae as an example. In Saccharomyces cerevisiae, the transcription units encoding ribosomal RNA genes are repeated 200 times on the chromosome 12.
This shows one transcription unit of them. This transcript is called 35S ribosomal RNA. AAA. Within one transcription unit, there are genes for 18S ribosomal RNA, 5.8S ribosomal RNA, 25S ribosomal RNA, and 5S ribosomal RNA genes. The 18S, 5.8S, and 25S ribosomal RNA genes are transcribed in this direction by Polymerase I. On the other hand, the 5S ribosomal RNA is transcribed in this direction by Polymerase III.
The 35S ribosomal RNA transcript is subsequently cleaved by RNase, resulting in the final products of 18S, 5.8S, and 26S ribosomal RNAs.
13. Assembly of eukaryotic ribosome in nucleus
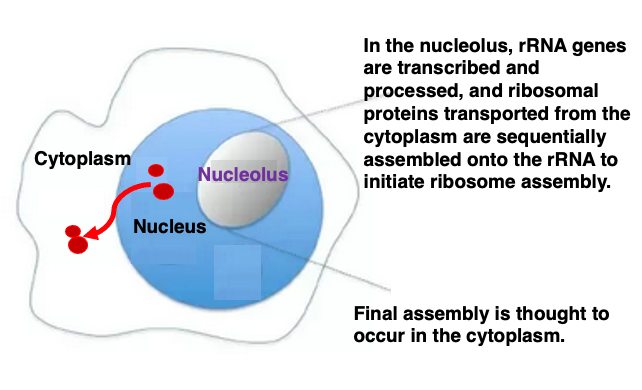
I will then explain on the assembly of eukaryotic ribosome. In eukaryotes, the genes coding for ribosomal RNA are transcribed in the nucleolus region within the nucleus.
Four types of ribosomal RNAs are processed and assembled in the nucleolus region, while ribosomal proteins are transported from the cytoplasm into the nucleus, sequentially attaching to the folding ribosomal RNAs to form their three-dimensional structures.
Partially assembled ribosome particles are then exported through the nuclear membrane into the cytoplasm, where final fine-tuning and complete assembly takes place. Therefore, the nucleolus is considered the site of ribosome assembly.
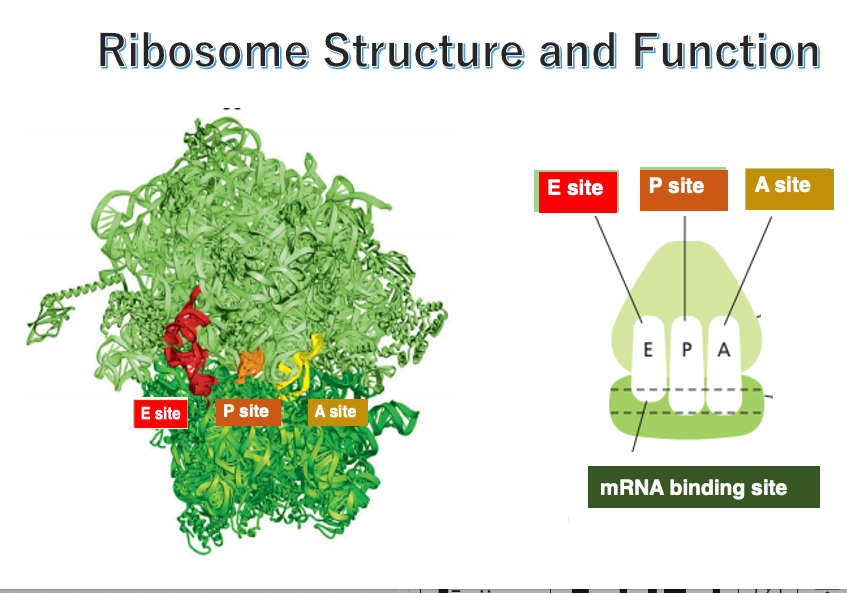
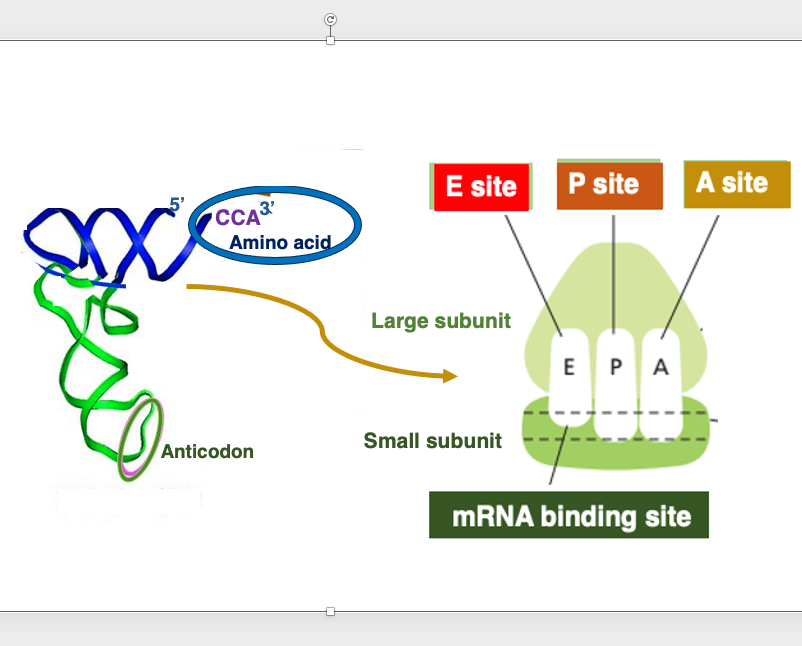
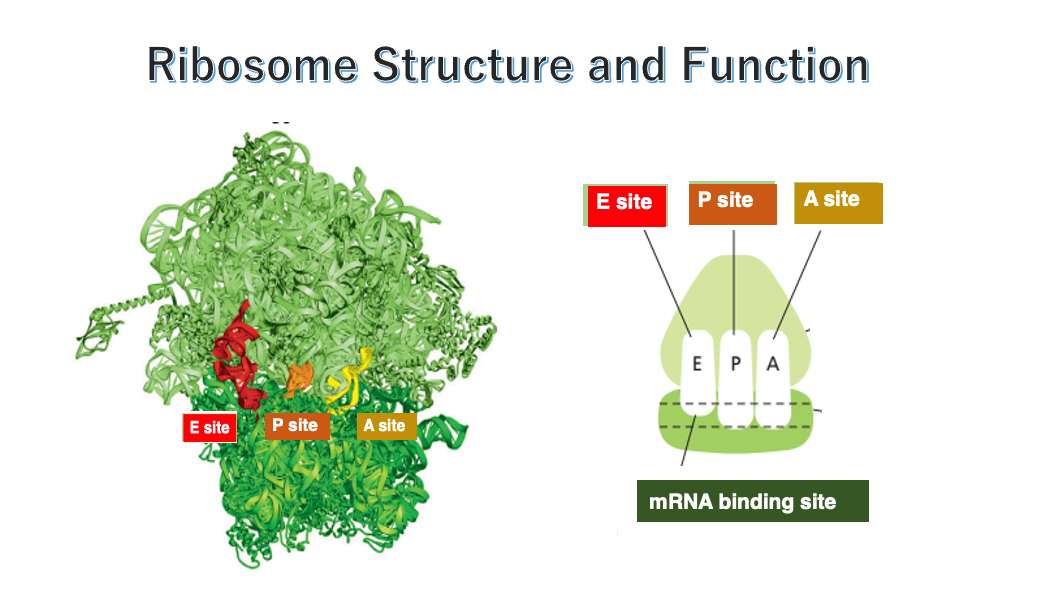
14. Internal cavities in the ribosome
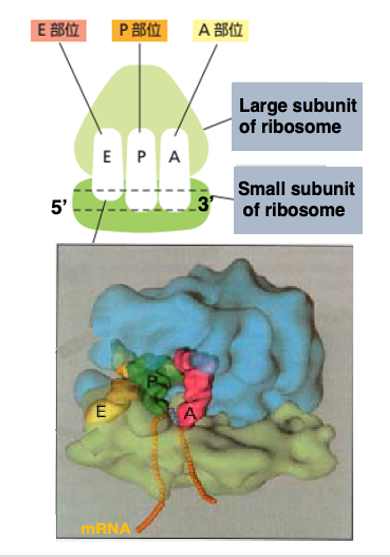
This figure shows an image of the internal space in the ribosome. In both eukaryotes and prokaryotes, the ribosome contains three cavities.
Each one of them can accommodate a single transfer RNA. These sites are named E-site, P-site, and A-site, respectively, in the direction from the 5’ to the 3’ end of the messenger RNA.
The E-site, short for Exit site, is where the deacylated tRNA exits the ribosome after the polypeptide chain is released. The P-site holds the tRNA carrying the growing polypeptide chain at its 3’ end. The A-site is where the incoming aminoacyl-tRNA binds, allowing the addition of the next amino acid to the polypeptide chain.
The small subunit captures the messenger RNA in this manner.
15. Action of ribosomes and tRNA in translation
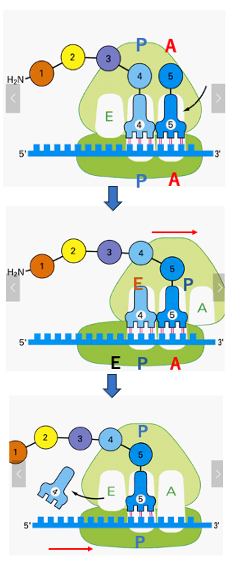
The following is an explanation of the movement of the ribosome and transfer RNA, during the peptide chain elongation reaction. In this figure, transfer RNA with a peptide consisting of four amino acids is in the P site. On the right side of it, the transfer RNA with the fifth amino acid is about to invade. When the pairing of the codon and the anticodon of the fifth transfer RNA is confirmed to be correct, the large subunit of the ribosome is shifted by three bases in the 3 ‘-direction. At the same time, the peptide chain attached to the fourth transfer RNA binds to the amino acid carried by the fifth transfer RNA. In other words, peptide chain transfer occurs.
Looking at the position of the transfer RNA immediately after the peptide transfer, now the 4th transfer RNA is in the E site of the large subunit, but it is in the P site of the small subunit, because the small subunit is not moving yet. The fifth-transfer RNA that received the peptide chain is located in the cavity created at the P site of the large subunit and the A site of the small subunit. Thus, fifth-transfer RNA is contained in a hybrid site composed of different sites in the large and small subunits.
Next, the small subunit is shifted in the 3’ direction by 3 bases. In accordance with this movement of the small subunit, the number 4 transfer RNA leaves the E site and migrates into the cytoplasm. The number 5 transfer RNA, which has a peptide chain, is located in the regular P site. Now, the number 6 transfer RNA can invade the A-site, and then peptide strand transfer from the number 5 transfer to the number 6 transfer RNA occurs again.
By repeating this sequence of reactions, the peptide chain is extended one amino acid residue at a time.
16. Elongation reaction of peptide chain in chemistry
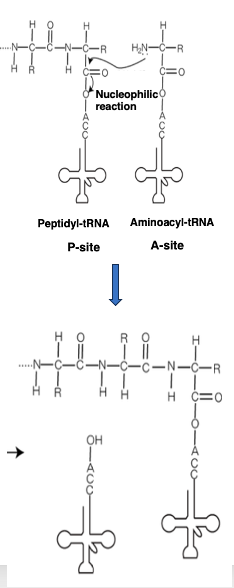
From an organic chemistry perspective, the peptide chain elongation reaction occurring in the ribosome can be described as follows:
The transfer RNA located at the P-site has the peptide chain attached to its 3‘-end. On the other hand, an aminoacyl-transfer RNA carrying an amino acid enters the A-site.
The amino group of this transfer RNA performs a nucleophilic attack on the carboxyl group of the peptide chain, resulting in the formation of a peptide elongated by one amino acid residue.
17. rRNA is a catalytically active RNA = Ribozyme
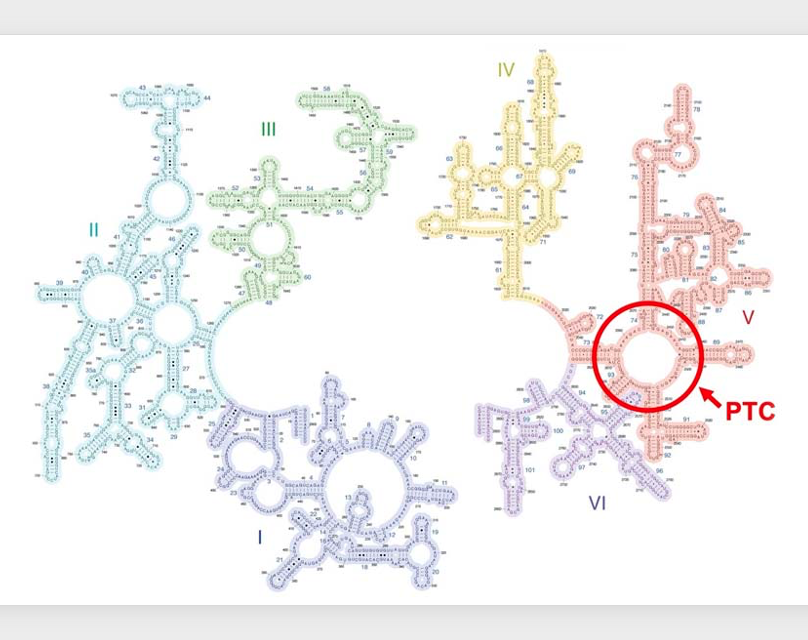
The peptide transfer reaction, I just explained is carried out extremely rapidly even at room temperature due to the catalytic activity possessed by the ribosome. This catalytic activity resides in a region of the 23S ribosomal RNA within the large subunit, known as the Peptidyl Transferase Center. In other words, the 23S ribosomal RNA can be considered a biomolecule possessing catalytic activity.
As you know, most biomolecules with catalytic activity are proteins, recently, it discovered that RNA molecules can also exhibit catalytic activity in cell. Such RNA molecules that function as enzymes are specifically referred to as ribozymes. Apart from ribosomal RNA, a representative example of a ribozyme is the spliceosomal RNA.
In cases where protein-RNA complexes carry out complex biochemical reactions, the active site often resides in the RNA component rather than the protein. This is due to the fact that in the early stages of life, it was RNA molecules, not proteins, that possessed enzymatic activity, , and proteins were molecules that assisted RNA molecules to stably adopt the conformation necessary for enzymatic activity.
コメント