This lecture can be viewed on YouTube site: Life Scinence Lectures for you
https://youtu.be/SoPHtxR7T2Q
Lecture contents
- Barbara McClintock’s proposal of the necessity of mobile genetic elements in the genome
- Proportions of various transposons in the human genome
- Classification of transposons: DNA transposons, Retrotransposons
- Classification within Retrotransposons: LTR-type Retrotransposons and Non-LTR-type Retrotransposons (LINE, SINE)
- Transposition mechanism of DNA transposons
- Transposition mechanism of LTR-type Retrotransposons
- Transposition mechanism of LINEs
- Transposition mechanism of SINEs
9. Mechanisms for suppressing transposition
Key words: Barbara McClintock, DNA transposon, Retrotransposon, LTR- Retrotransposon, Non-LTR Retrotransposon, LINE, SINE, transposase, gag, pol, env, integrase, VLP, telomerase, copy and paste, target-primed reverse transcription, endonuclease
1. Necessity of Mobile Genetic Elements on the Genome
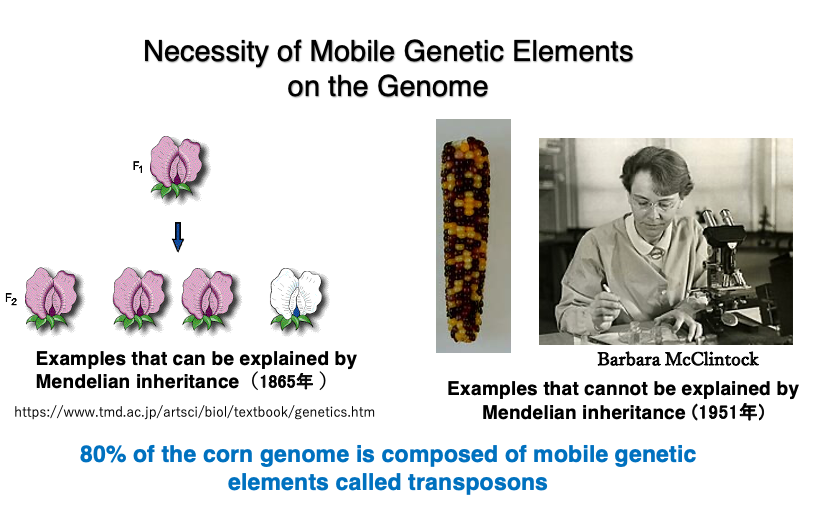
Mendel hypothed the existence of genes on genome that determine the flower colors and other traits in plants. This allowed him to successfully explain the segregation ratios of flower colors resulting from crossbreeding, leading to the publication of his paper in 1865. The Mendelian genes were assumed to be stably positioned on chromosomes and constantly expressed.
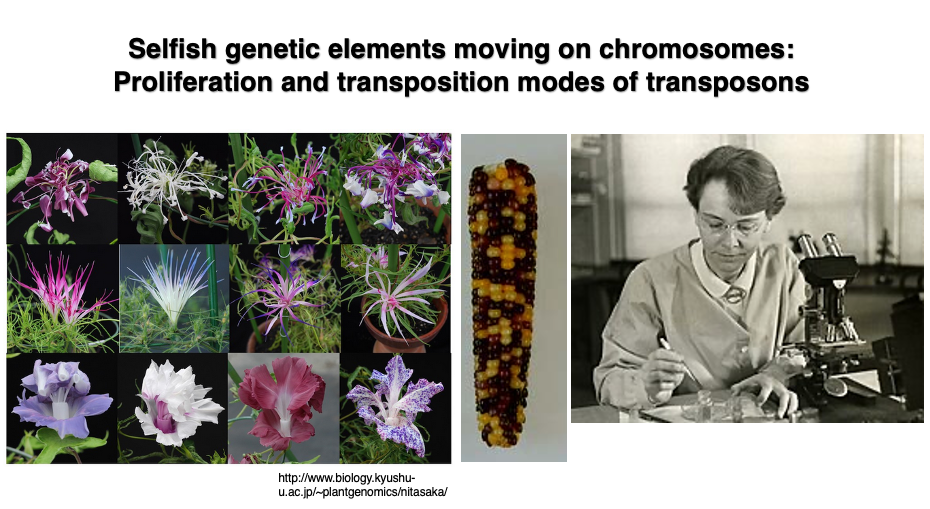
About 90 years later, in 1951, Barbara McClintock discovered phenomena that could only be explained by the presence of mobile genetic elements within the genome while conducting experiments to create a genetic map of maize. She named these mobile elements “transposons.” As shown in the figure, the maize kernels are a mixture of purple-red and yellow. McClintock discovered that such changes in seed color could not be explained by Mendelian genetics.
The existence of mobile elements was later confirmed at the DNA sequence level. For this work, she was awarded the Nobel Prize in 1983. Today, analysis of whole genome sequence data has revealed that 80% of the maize genome is composed of transposons and their remnants.
2. Mutations caused by transposon transposition in Japanese morning glory
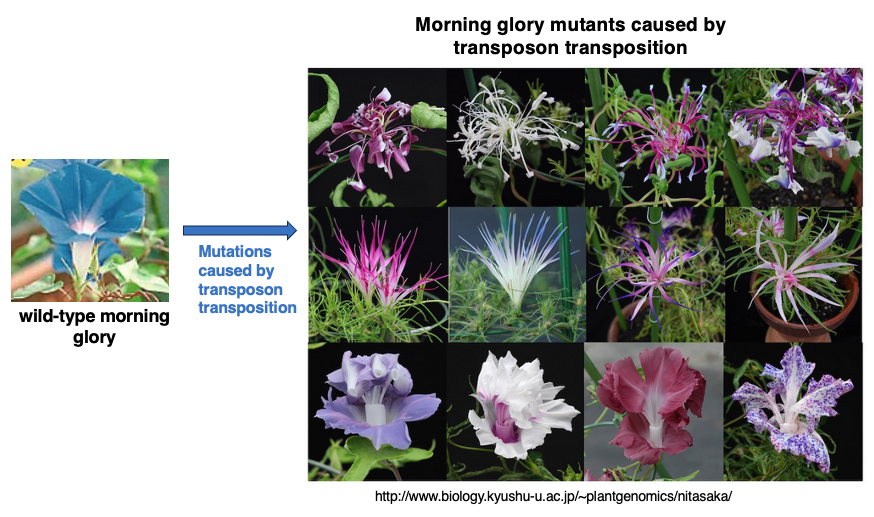
Various mutants are observed in Japanese morning glory. Many of these mutations are not caused by base mutations that occur during genome DNA replication, but rather by transposons jumping across the genome, increasing their copy numbers, and thereby disrupting existing genes or altering their expression.
From a human perspective, this behavior of transposons may seem like a selfish factor whose sole purpose is to increase its own copies. This lecture will explain the transposition mechanisms of transposons and how they increase their copy numbers.
3. Proportions of Various Transposons in the Human Genome
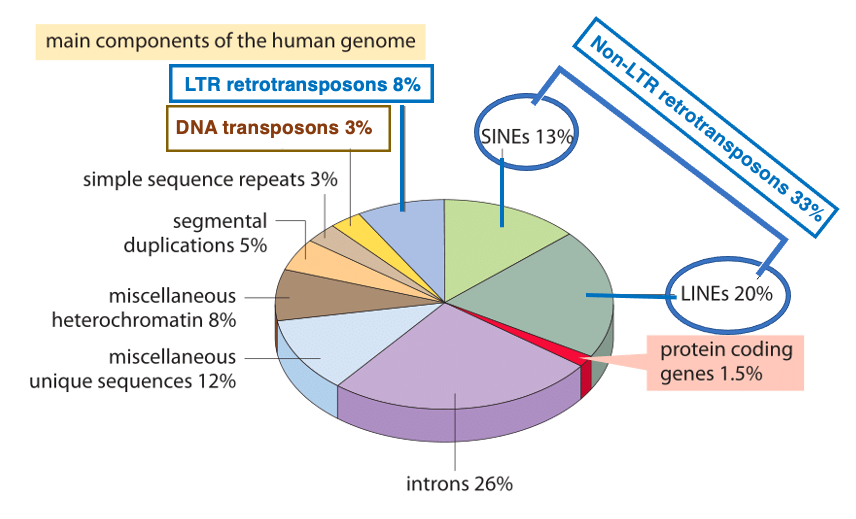
This pie chart shows the proportions of elements that make up the human genome. First, when all the exon parts of protein-coding genes are combined, they account for 1.5% of the genome. On the other hand, the total of introns amounts to 26%. As exceptions, there are genes without introns, such as histones, G-proteins, and some transcription factors. However, other than these, protein-coding genes have multiple introns.
There are two types of transposons: DNA transposons and retrotransposons. Retrotransposons are further divided into those with Long Terminal Repeats (LTRs) of 200-600 bp at both ends, and those without LTRs (Non-LTR). DNA transposons make up 3% of the genome, while LTR-type retrotransposons account for 8%. Non-LTR types are further classified into SINEs and LINEs, which together comprise 33% of the genome. When we add the 8% of DNA transposons to the 41% of retrotransposons, we find that transposons occupy about half, or 49%, of human genome.
However, many transposons have currently lost their transposition activity. In humans, there are no DNA transposons with transposition activity at present. Moreover, active retrotransposons are limited to three types: LINE-1 (L1), Alu, and SVA.
4. Proportion of transposons in genome across various species
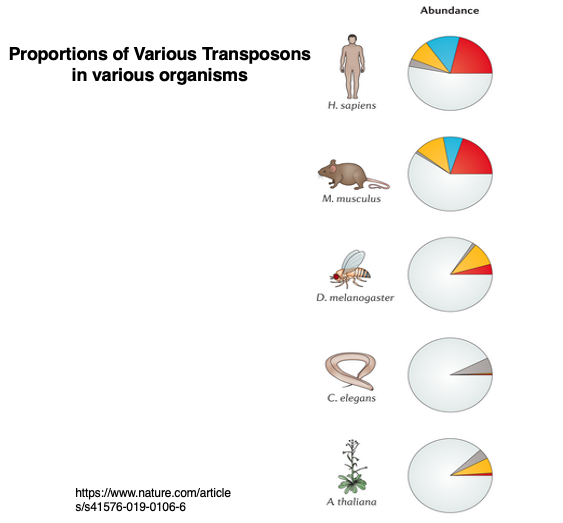
This pie chart shows the proportion of transposons in the genomes of various species. As you can see, the ratio of transposons varies greatly among different organisms. Humans can be considered a species with a relatively high proportion of transposons.
While Drosophila melanogaster has 3-5% transposons, Drosophila simulans has 11-13%, demonstrating that even closely related species can have significantly different proportions of transposons. The question of how active transposons emerge and become inactive is an intriguing topic. It is believed that at least some transposons can move across species boundaries.
5. DNA transposons and Retrotransposons
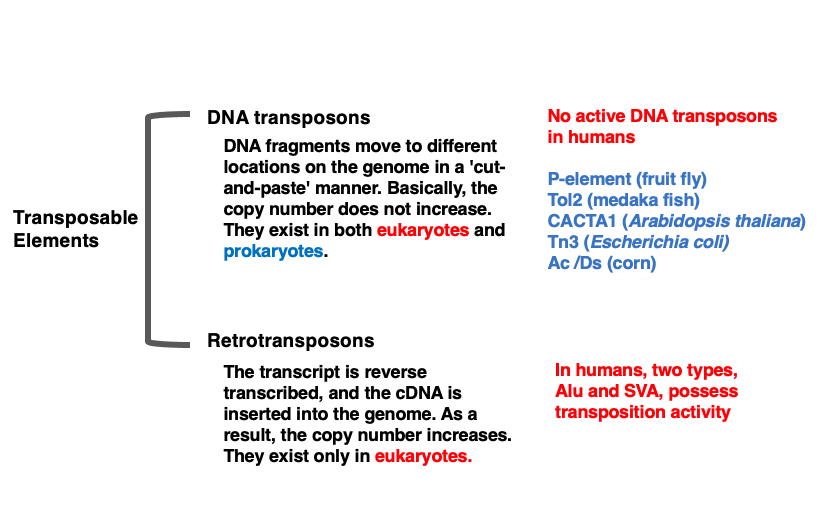
Transposons are broadly classified into two categories based on their mode of transposition: DNA transposons and Retrotransposons. DNA transposons are genetic elements that are excised from the genome as DNA and inserted into a different location through a ‘cut-and-paste’ mechanism.
Since this is a ‘cut-and-paste’ form of transposition, it is thought that the copy number within the genome does not easily increase. DNA transposons are believed to share a common ancestor with DNA viruses.
Currently, there are no DNA transposons with transposition activity in human. However, remnants of what are believed to have once been active transposons occupy about 3% of the human genome. On the other hand, DNA transposons with active transposition ability are known to exist in other eukaryotes and prokaryotes.Retrotransposons are transposons that increase their copy number through RNA. The transcribed mRNA is converted into cDNA by reverse transcriptase, which is then integrated into the genome. Since mRNA, the transcription molecule, can be produced in multiple copies, there is a high possibility that numerous cDNA molecules will be created, leading to the characteristic of easily increasing copy numbers. Retrotransposons exist only in eukaryotes and are not found in prokaryotes.
6. Classification of Retrotransposons
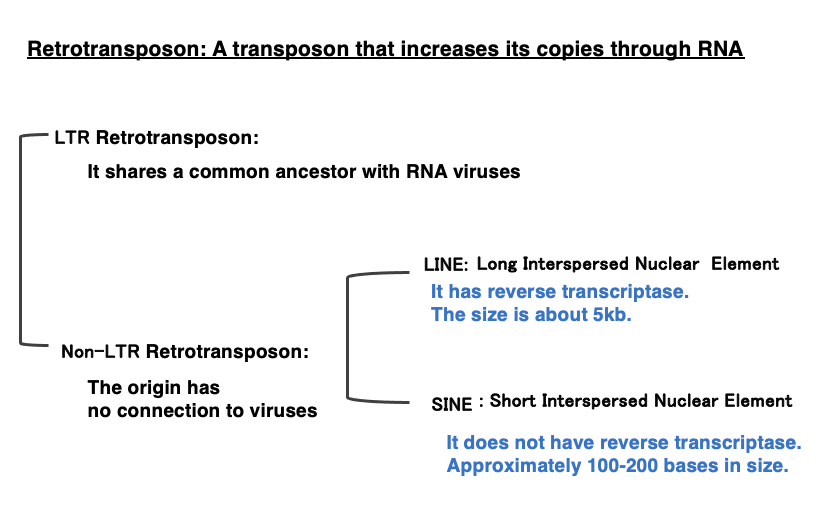
Retrotransposons are broadly categorized into two types: LTR retrotransposons and Non-LTR retrotransposons. LTR retrotransposons are believed to share a common ancestor with RNA viruses that propagate through reverse transcriptase. Therefore, they can also be referred to as RNA virus-type retrotransposons.
On the other hand, Non-LTR retrotransposons do not share ancestral commonality with RNA viruses. Non-LTR retrotransposons are further divided into two categories:
Short Interspersed Nuclear Elements (SINEs): These are about 100-400 bases in length and do not encode reverse transcriptase.
Long Interspersed Nuclear Elements (LINEs): These are approximately 5,000 base pairs in size and encode their own reverse transcriptase internally.
7. DNA Transposon’s Transposition Mechanism
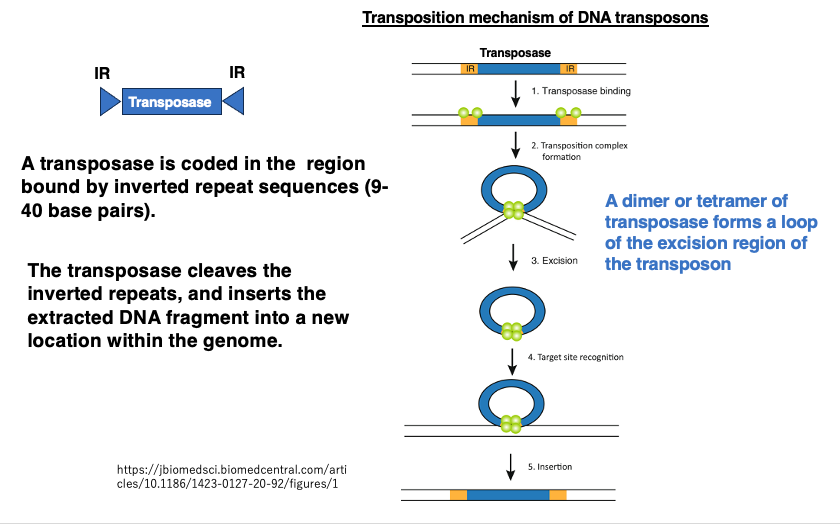
First, let’s explain the transposition mechanism of DNA transposons. At both ends of this transposon, there are Inverted Repeat sequences consisting of 9-40 base pairs. The region enclosed by these Inverted Repeat sequences codes for Transposase.
Transposase attaches to the Inverted Repeat sequences and pulls together the Inverted Repeat sequences at both ends, forming a loop and then cleaving it. The complex of the cleaved DNA transposon and Transposase then cuts a new site in the genome and inserts the excised DNA fragment into that location. If the DNA transposon is inserted within a gene, it can cause gene disruption. Even when inserted into an intron, it may interfere with normal splicing.
8. DNA transposon Tn3 in Escherichia coli
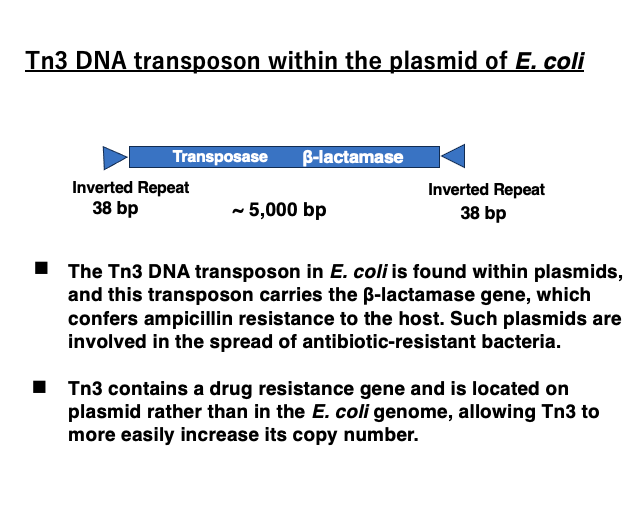
The DNA transposon Tn3 is a transposon of nearly 5,000 base pairs. It has 38-base pair inverted repeats at both ends. Internally, it encodes a transposase as well as a β-lactamase gene. As a result, it can break down antibiotics such as ampicillin, making the host resistant to ampicillin.
Furthermore, Tn3 is inserted into plasmids rather than the genome, allowing ampicillin resistance to spread as the plasmid is passed on to other bacterial cells. From a different perspective, by incorporating an antibiotic resistance gene and being located within plasmids, Tn3 has acquired advantageous conditions for increasing its own copy number.
9. Classification Retrotransposons
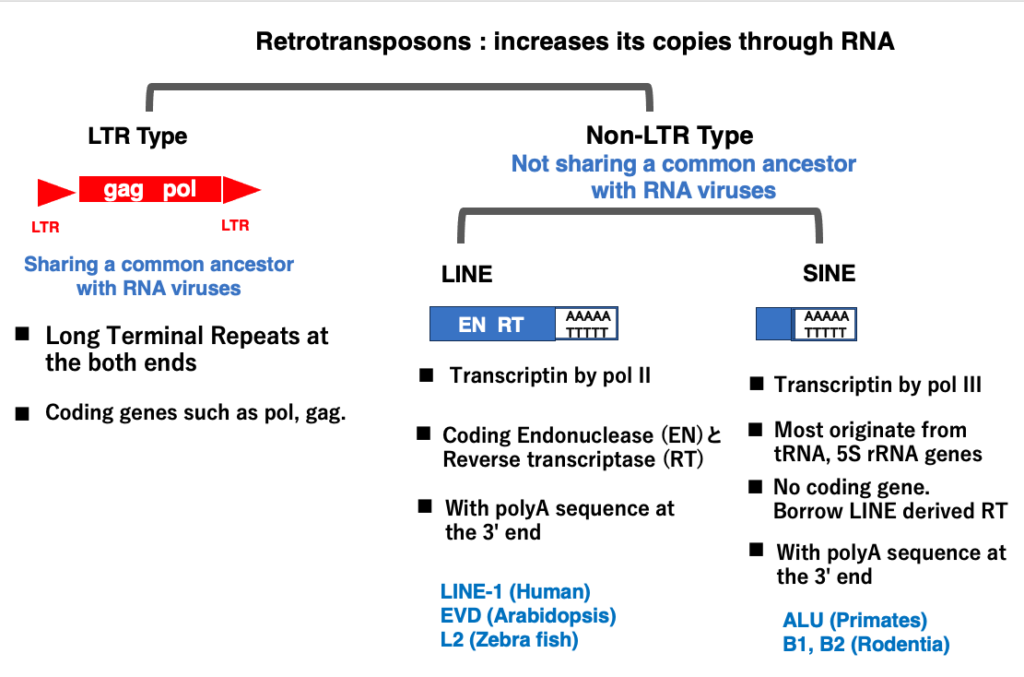
Retrotransposons are a class of transposable elements that replicate through an RNA intermediate. The coding region of a retrotransposon is transcribed by RNA polymerase and then reverse transcribed by reverse transcriptase. The resulting cDNA is inserted into a new location in the chromosome. Consequently, if multiple transcripts are produced, multiple cDNA molecules are created, leading to a rapid increase in copy number within the genome. In other words, retrotransposons can be described as transposons that multiply via RNA.
LTR retrotransposons, which have Long Terminal Repeats (LTRs) at both ends, share a common origin with RNA viruses. It’s easier to understand them as RNA viruses that have lost the envelope protein gene necessary for escaping the cell after invading it.
On the other hand, Non-LTR retrotransposons are thought to have origins unrelated to RNA viruses. Non-LTR retrotransposons are further classified into LINEs and SINEs. While LINEs and SINEs share the same transposition mechanism, their molecular origins differ.
LINEs are transcribed by RNA polymerase II and contain internal coding regions for proteins necessary for transposition. SINEs, however, lack these coding regions and originate from small RNA genes like tRNA or 5S rRNA, which are transcribed by RNA polymerase III.
Both LINEs and SINEs contain a 20-50 base pair 3′-AAAA-5’/5′-TTTTT-3′ DNA sequence in their 3′ region. Consequently, their transcripts have a poly A RNA sequence at their 3′ end. This poly A sequence plays a crucial role in the reverse transcription and insertion of Non-LTR retrotransposons into new genomic locations.
10. On the transposition mechanism of Retrotransposons with LTR
I will explain the transposition mechanism of Retrotransposons with LTRs. These Retrotransposons share a common ancestor with RNA viruses.
11. Genome structure of retroviruses that replicate using reverse transcriptase

LTR-type retrotransposons share a common ancestor with viruses, so let’s first review retroviruses that replicate using reverse transcriptase. This is a diagram of the structure and genome composition of human HIV-1.
This type of RNA virus has long inverted repeats (LTRs) at both ends, which act as promoters for RNA polymerase II. Internally, they commonly contain three genes: gag, pol, and env.
The Gag gene encodes capsid proteins. The product of the Pol gene is cleaved by proteinase and becomes proteins with Reverse transcriptase, RNase H, and Integrase activities. The Env gene encodes envelope proteins, which form the outermost shell of the virus.
12. Genome structure of LTR retrotransposons
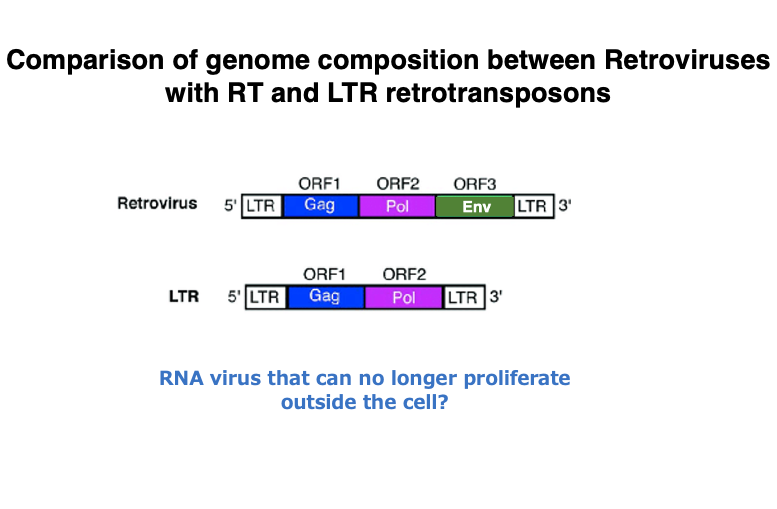
This is a comparison of the genome structures of retroviruses and LTR retrotransposons, both of which possess reverse transcriptase.
In transposons with LTRs, the gene that produces the envelope protein found in retroviruses is absent. It may be easier to understand LTR retrotransposons if you consider them as molecules that originated from retroviruses that invaded the genome. During their latency in cells, these retroviruses lost their envelope gene, thereby losing the ability to proliferate outside the cell.
13. Transcription and Transposition of LTR- retrotransposons
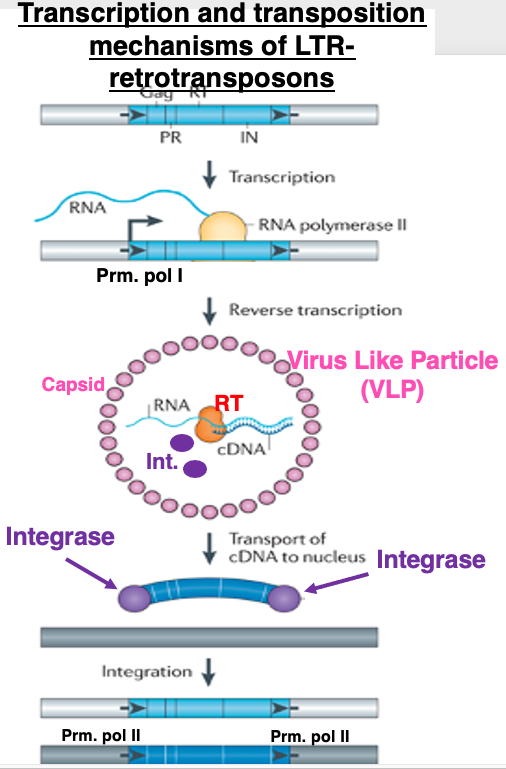
I will explain the outline of transcription and transposition of retrotransposons with LTRs. Transcription is carried out by RNA polymerase II from the promoter located in the Long Terminal Repeat on the 5′ side. The mRNA moves to the cytoplasm and is translated by the host’s ribosomes.
Virus Like Particles (VLPs) are formed when capsid proteins encoded by the gag gene assemble into particles that incorporate mRNA, Reverse transcriptase and Integrase derived from the pol gene. Reverse transcription occurs within this VLP, ultimately producing double-stranded cDNA.
The double-stranded cDNA, with integrase attached to its ends, moves into the nucleus. Through the action of integrase, the cDNA is inserted into the genome. The insertion site is thought to be almost random. If inserted into a gene, it can cause gene disruption. Also, since the LTRs at both ends have RNA polymerase II promoter activity, they may affect the transcription of nearby host genes.
The process of reverse transcription of this mRNA is quite complex. While the mRNA is produced from the middle of the 5′-UTR to the end of the 3′-UTR, by utilizing the 3′-UTR sequence, the resulting cDNA has complete LTRs placed at both the 5′ and 3′ ends, identical to the original. Therefore, even in the transposed copy DNA, the promoter activity is maintained, allowing it to transcribe its own genes regardless of where it is inserted in the genome.
14. Detailed mechanism of LTR retrotransposon transposition
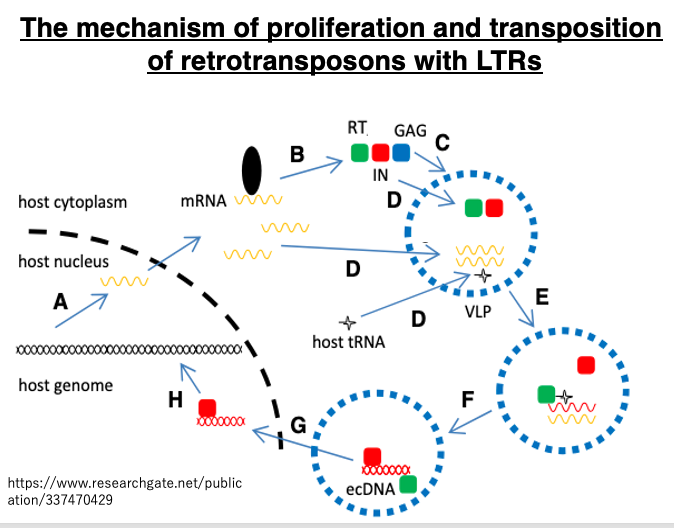
Let’s review the transposition of LTR retrotransposons using a different diagram. In the nucleus, the transposon is transcribed by pol II from the LTR. The mRNA that moves to the cytoplasm is translated, producing capsid proteins (blue) from the gag region, and Reverse transcriptase (green) and integrase (red) from the pol region. The capsid proteins assemble to form a Virus Like Particle (VLP). Inside this particle, the mRNA is reverse transcribed, ultimately becoming double-stranded cDNA.
Host tRNA is used as a primer for this reverse transcription reaction. The cDNA and integrase move into the nucleus. The cDNA is inserted into the genome by integrase. This LTR-retrotransposon cDNA retains an intact 5′-LTR containing promoter sequences, so it can transpose further.
15. LINE Transposition Mechanism
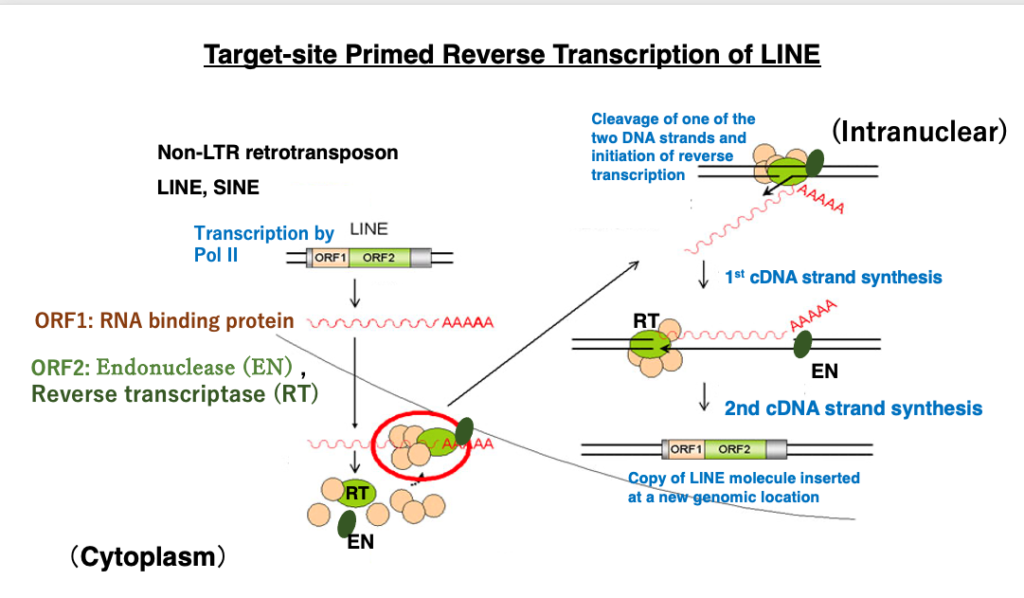
Next, I will explain the transposition of LINE, a type of Non-LTR retrotransposon. While LTR retrotransposons are closely related to RNA viruses, Non-LTR retrotransposons also replicate themselves through RNA reverse transcription but do not share a common ancestor with RNA viruses, except for the reverse transcriptase. Non-LTR retrotransposons contain two genes encoding ORF1 and ORF2. ORF1 encodes an RNA binding protein that attaches to its own mRNA. ORF2 encodes an endonuclease (EN) with weak specificity and a reverse transcriptase (RT).Transcription is carried out by RNA polymerase II from a promoter within the 5′-UTR, and translation is performed by the host ribosomes. The translation products – RNA binding protein, endonuclease, and reverse transcriptase – attach to their own mRNA, which is then returned to the nucleus.
In the nucleus, when the endonuclease cleaves the genome, reverse transcription begins from the 3′-OH of the DNA cleavage site, using the retrotransposon mRNA as a template. Eventually, the double-stranded cDNA is inserted at the genome cleavage site. This mechanism is called target-primed reverse transcription transposition.
16. Details of the LINE-1 transposition mechanism
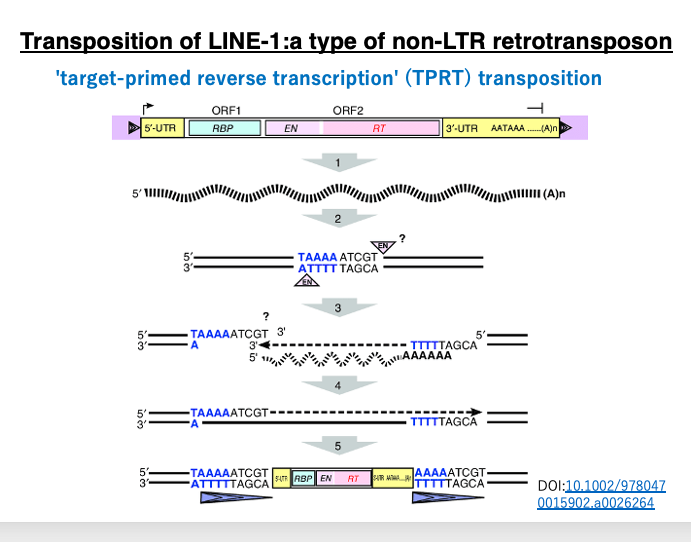
Let’s examine the LINE-1 target-primed reverse transcription transposition in more detail using a different diagram. Transcription begins from the promoter in the 5′-UTR region by RNA polymerase II, transcribing the ORF1 (which encodes an RNA binding protein), ORF2 (which encodes endonuclease and reverse transcriptase), and the 3′-UTR including the poly A sequence.
Typically, RNA polymerase II initiates transcription 20-30 bases downstream of the TATA box in the promoter. Therefore, mRNA transcribed by pol II usually does not contain the promoter sequence. However, the LINE pol II promoter is unique. It’s a type of internal promoter where transcription begins upstream of the promoter sequence, so the resulting mRNA includes the promoter sequence. Consequently, the cDNA also contains the promoter, allowing the transposed copy to transcribe its own gene again.
LINE-1 endonuclease has a weak base specificity, recognizing the 5′-TTTA-3′ sequence and cleaving between T and A (5′-TTT|A-3′). Unlike restriction enzymes, the recognition sequence doesn’t need symmetry, so the cleavage point on the other strand is unclear but is thought to be about 5 bases upstream of the 5′-TTT|A-3′ sequence.
In this diagram, the cleaved DNA has a 9-base 3′ overhang. The poly A at the 3′ end of the transcribed LINE-1 forms hydrogen bonds with the TTT on the 3′ overhang of the cleaved genomic DNA. Using the attached LINE-1 mRNA as a template, reverse transcription occurs as the DNA 3′ end extends.
Eventually, the LINE-1 mRNA becomes a complete double-stranded DNA. At this time, the 9-base 3′ overhang of the genome that was initially cleaved is also repaired to double-stranded DNA. As a result, both ends of the LINE-1 copy molecule are flanked by this 9 bp inverted repeat (shown as elongated triangles in the figure).
17. SINE transposition mechanism
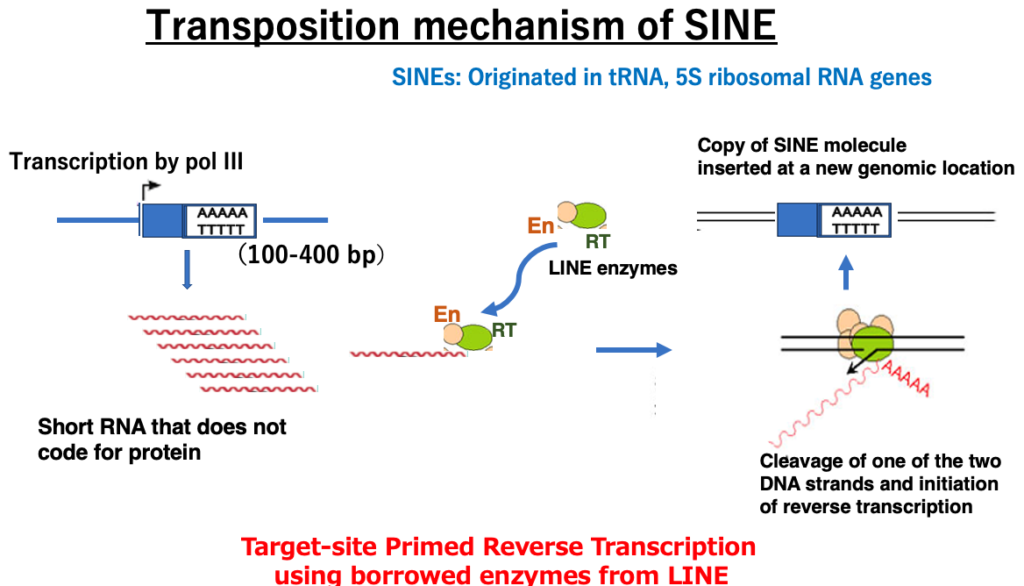
Next, I will explain the transposition mechanism of SINEs, which are another type of retrotransposon without long terminal repeats like LINEs, but are shorter molecules. Most SINEs originate from tRNA or 5S rRNA genes transcribed by RNA polymerase III. As a result, they are short (100-400 bp) and do not contain ORFs internally, but unlike their tRNA or 5S rRNA origins, they have a short poly A sequence at the 3′ end of the DNA (5′-AAAAA-3’/3’TTTTT-5′).
This feature is the same as LINEs. The SINE transcript is joined by LINE-derived endonuclease and reverse transcriptase in the cell and translocates to the nucleus. Borrowing LINE-derived enzymes, SINEs insert copies of themselves into new positions in the genome through ‘target-primed reverse transcription’ (TPRT) transposition, just like LINEs. In terms of creating copies of themselves in the genome by borrowing LINE enzymes, SINEs can be considered more parasitic molecules than LINEs.
18. RNA polymerase III promoter
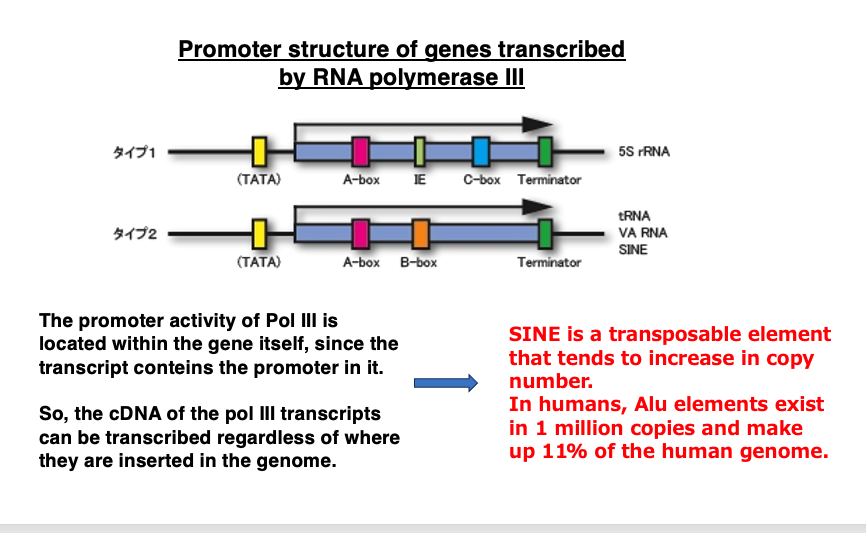
As shown in this figure, the promoter for RNA polymerase III is contained within the gene it transcribes. Therefore, even if the transcript is converted to DNA, it can be transcribed again.
Additionally, since it’s a short molecule, unlike LINE, it’s less likely to have its reverse transcription interrupted midway. For these reasons, it’s considered to be a molecule that can easily proliferate. In fact, Alu, a type of active SINE molecule in humans, exists in 1.2 million copies.
19. Copy number of transposable elements in the human genome
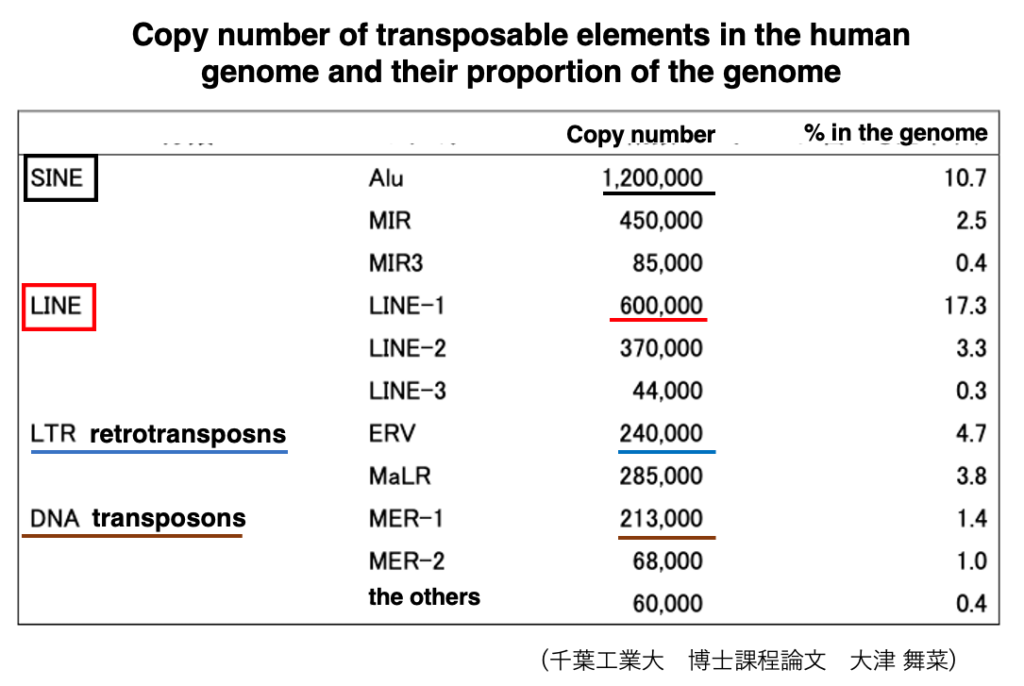
If we look more closely at the copy numbers of transposons within the human genome, it appears as shown in this table. It shows in human, SINE shave a stronger ability to increase their copies compared to other transposons.
The order of transposons from highest to lowest copy number is SINE, LINE, LTR-retrotransposon, and DNA transposon.
20. The close relationship between reverse transcriptase and telomerase
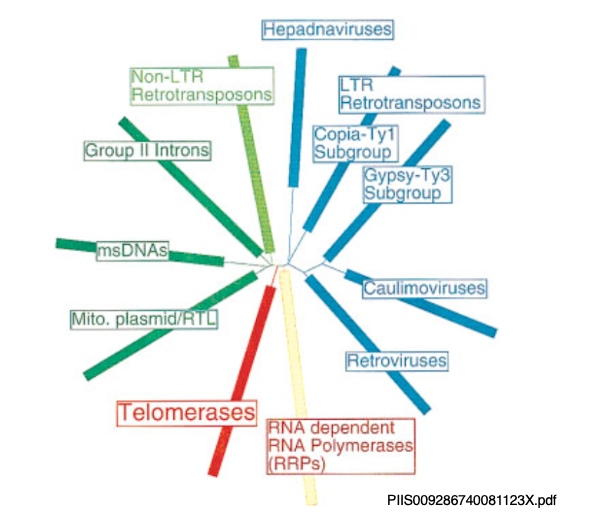
Whether LTR-type or Non-LTR-type, reverse transcriptase is essential for the transposition of retrotransposons. Reverse transcriptase is also crucial for the transposition of retroviruses and Group II introns.
Moreover, telomerase, which is involved in the extention of repetitive sequences at the ends of genomes, is also a type of reverse transcriptase. While their evolutionary relationships are unclear, telomerase is an enzyme essential for the emergence of eukaryotes with linear genomes. The fact that it is closely related to rverse transcriptase is very intriguing.
21. Mechanisms to suppress transposon transposition activity
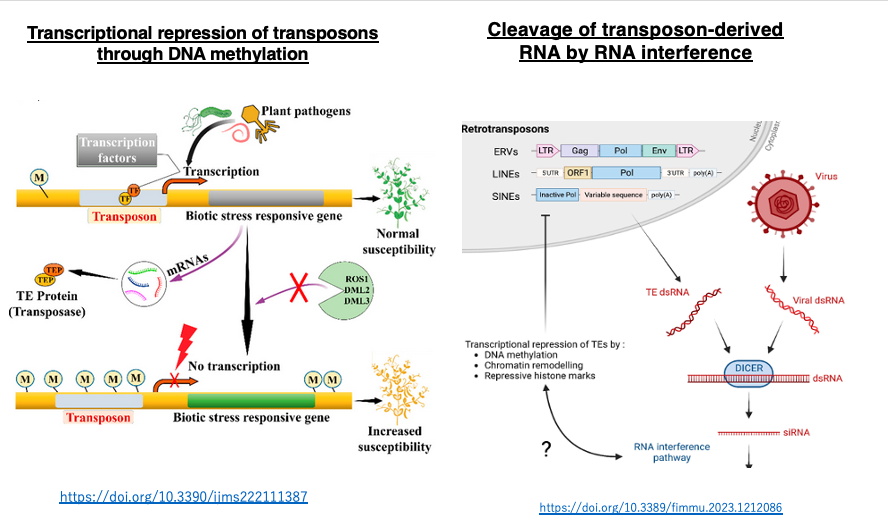
When transposons actively transpose in somatic cells, they can induce gene disruption and changes in host gene transcription levels, which in most cases have harmful effects on the organism. On the other hand, transposon transposition in germ cells can be a driving force for evolution.
In eukaryotes, long-term stable transcriptional suppression mechanisms have developed to inhibit transposon transposition, including DNA methylation of transposon-coding sequences, histone modifications, and RNA interference. These mechanisms are also repurposed for stable transcriptional repression of the host’s own intracellular genes, contributing to cell differentiation in multicellular organisms.
コメント