This lecture can be viewed on the following YouTube site : Life Science Lectures for You
https://youtu.be/oIxL8TS99i4
1. This lecture includes the following content
1. The amino acid attached to the initiator tRNA and the amino acids attached to elongator tRNAs.
2. Characteristics of the upstream sequence of the translation initiation AUG codon.
3. Formation of the 30S Small subunit complex at the start of translation.
4. Entry of aminoacyl-tRNA into the A-site and the shift of the Large subunit.
5. Shift of the Small subunit due to EF-G・GTP hydrolysis and resolution of the Hybrid site.
6. Recognition of the stop codon by Release Factors, release of the newly synthesized peptide chain, and separation of the large and small ribosomal subunits.
Key words: initiator tRNA, Elongator tRNA, fMet,Shine-Dalgarno sequence, IF-1, IF-2, IF-3, EF-Tu・GTP, EF-G・GTP,Speed and Accuracy of translation,Hybrid site, Release Factor, eRF1, RF-1, RF-2.
2. Formation of the translation initiation complex in the 30S Small subunit
In this lecture, I will explain the mechanism of bacterial translation step by step. First, I will explain the formation of the translation initiation complex in the 30S small ribosome. Let’s begin.
3. Two types of tRNAs can form stable pairs with AUG codons: In Eubacteria, Chloroplasts and Mitochondria
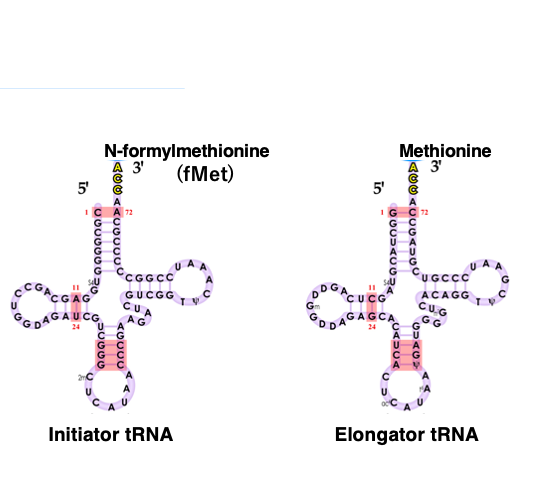
The start codon for translation is AUG, but the amino acid attached to the tRNA that pairs with this translation start AUG codon differs between eukaryotes and archaea, and bacteria and organelles.
Additionally, AUG is not exclusively a start codon and can also appear within the open reading frame. The tRNA that pairs with the translation start AUG codon is called initiator tRNA, while the tRNA that pairs with other AUG codons is called elongator tRNA.
In eubacteria, chloroplasts, and mitochondria, formylmethionine (fMet) is attached to the 3 prime end of the initiator tRNA. On the other hand, methionine is attached to the elongator tRNA.
4. Two types of tRNAs can form stable pairs with AUG codons: In Eukaryotes and Archaea
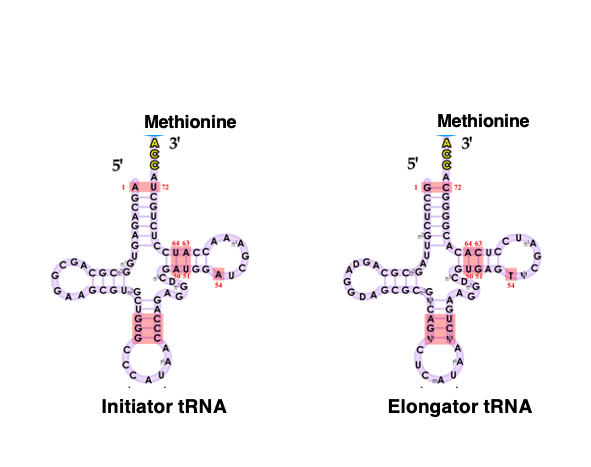
On the other hand, in eukaryotes and archaea, methionine, rather than formylmethionine, is attached to the initiator tRNA corresponding to the translation start AUG codon.
Similarly, methionine is also attached to the elongator tRNA.
5. Required conditions for the AUG codon to be a translation initiation codon
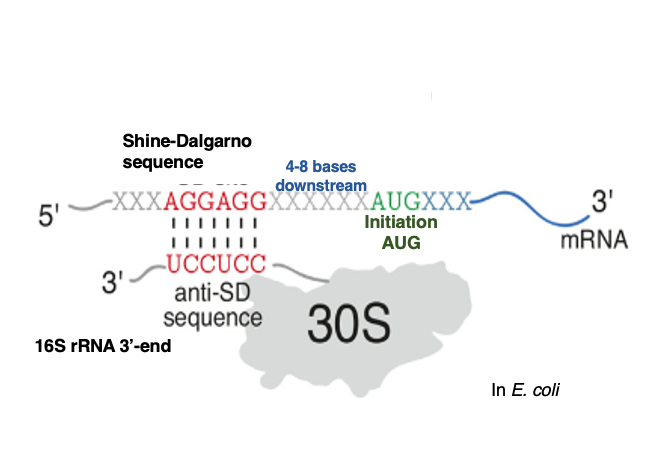
The AUG codon used for translation initiation has the following characteristics compared to AUGs that appear within the open reading frame. In the translation initiation AUG codon, a Shine-Dalgarno sequence is present 4 to 8 bases upstream of the AUG. The Shine-Dalgarno sequence refers to a sequence complementary to the 6-7 bases at the 3′ end of the 16S ribosomal RNA contained in the 30S ribosome. In the case of E. coli, the sequence 3′-AGGAGGU-5′ corresponds to this.
The Anti-Shine-Dalgarno sequence, located near the 3′ end of 16S ribosomal RNA, is thought to stabilize the attachment of mRNA to the ribosomal small subunit by forming hydrogen bonds with the Shine-Dalgarno sequence on the messenger RNA. On the other hand, such Shine-Dalgarno sequences are not found upstream of AUG codons that appear in the middle of Open Reading Frames.
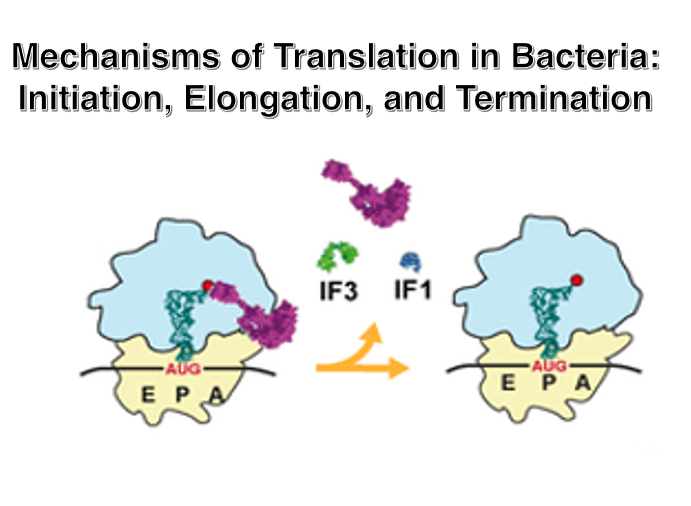
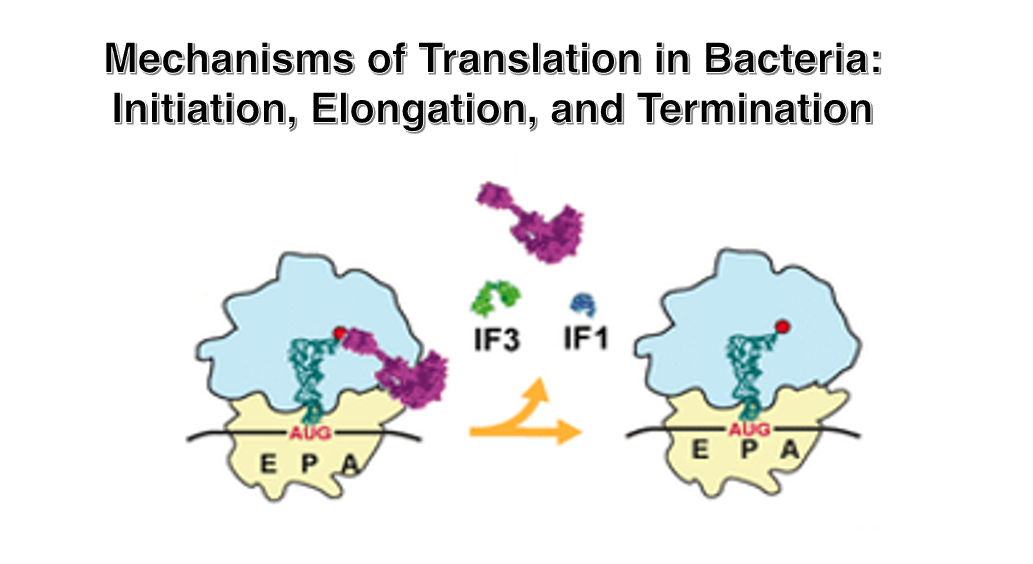
6. 30S Small ribosome translation initiation complex
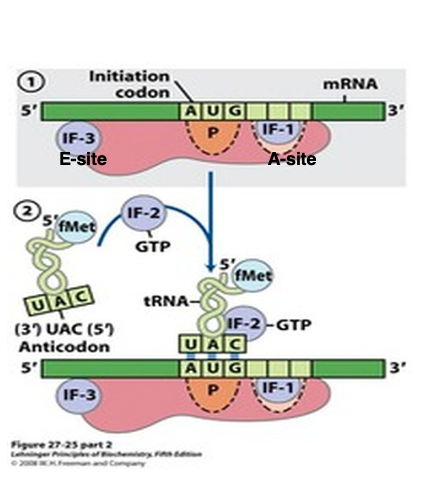
Prior to the initiation of translation, it is necessary for the small subunit to be dissociated from the large subunit.
Initiation Factor-3 (IF-3) attaches to the area near the anti-Shine-Dalgarno sequence of the 16S rRNA. The attachment of IF-3 to the small subunit facilitates the recruitment of mRNA. Additionally, through interactions between the Shine-Dalgarno sequence of the mRNA and the anti-Shine-Dalgarno sequence, the translation initiation AUG codon is positioned at the P-site of the small subunit. Furthermore, Initiation Factor-1 (IF-1), which attaches near the A-site, also promotes the binding of mRNA to the small subunit.
In this state, the initiator tRNA with attached formylmethionine (fMet) forms a ternary complex with the IF-2 • GTP complex, allowing it to stably bind to the translation initiation AUG codon.
7. Setup of initiator tRNA and Recruitment of the 50S large subunit
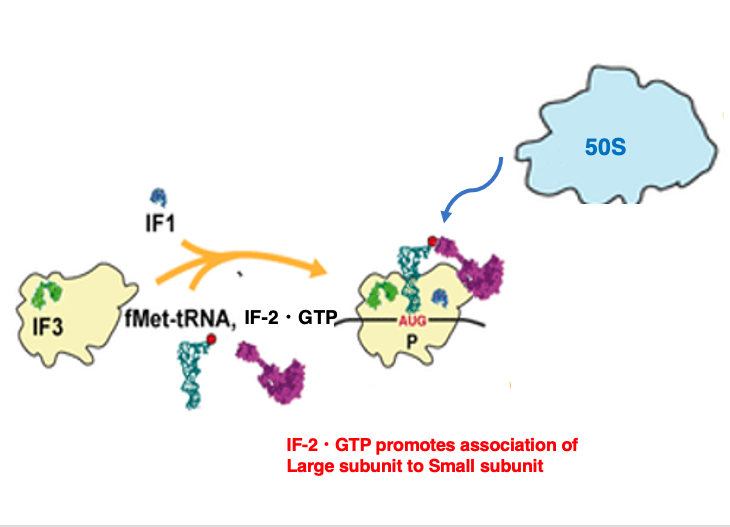
Once the setup of the translation initiation tRNA is completed, the large subunit is recruited onto the small subunit.
8. Formation of 70S ribosome complex
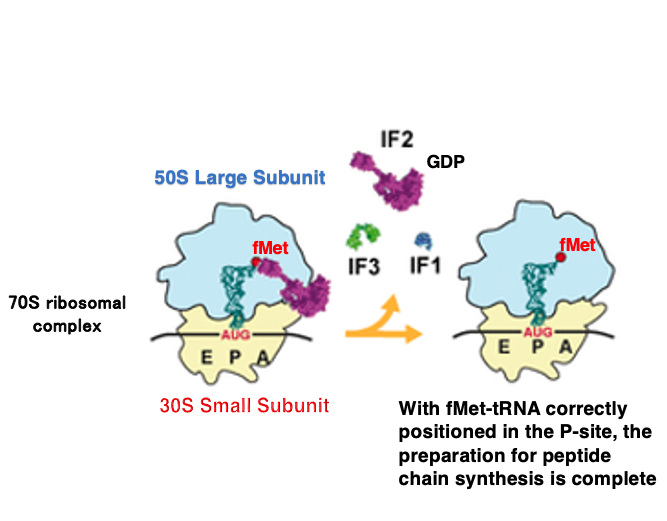
When the large subunit associates with the small subunit, the GTP in the IF-2 • GTP complex changes to GDP, causing the IF-2 • GDP complex and IF-1 to dissociate. Shortly after, IF-3 also dissociates.
Once this state is reached, the ribosome has completed its preparation for protein synthesis initiation and can accept an aminoacyl-tRNA into the empty A-site.
9. Ribosome setup process for translation initiation
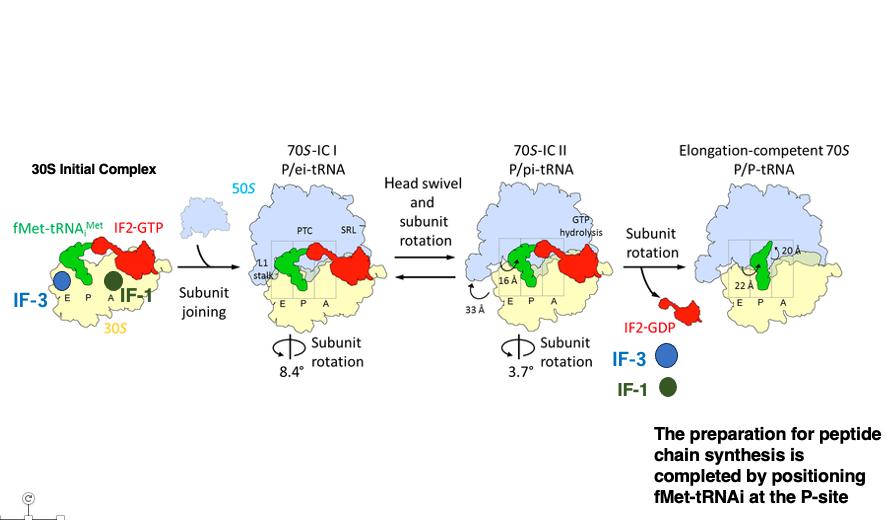
Let’s review the sequence of events using a different diagram. The translation start codon of mRNA is positioned at the P-site of the Small subunit, where the initiator tRNA carrying formylmethionine is recruited.
Finally, the Large subunit is brought in. The IF-1, IF-2, and IF-3 molecules used for setting up the translation initiation complex dissociate from the ribosome.
10. Access of aminoacyl tRNA to the A-site
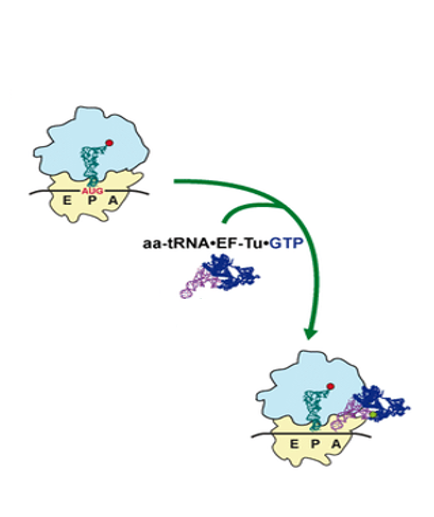
From here, I will explain the elongation reaction of the nascent peptide chain.
The initiator tRNA is in the P-site, and to its right, aminoacyl-tRNAs randomly access the A-site. The aminoacyl-tRNA is not transported to the A-site alone, but rather in a complex with Elongation Factor Tu・GTP.
Please be sure that it is IF-2・GTP used for the setting of fMet-initiator tRNA at the P-site AUG codon.
11. Reaction when the correct aminoacyl-tRNA enters the A-site
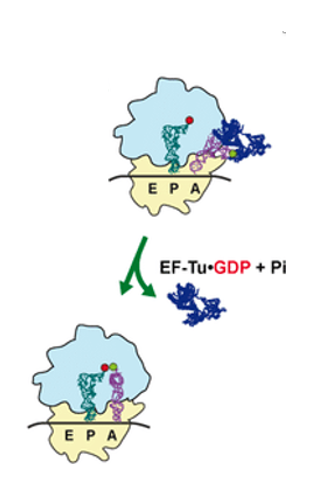
When an aminoacyl-tRNA enters the A-site, it is checked whether its anticodon forms stable hydrogen bonds with the codon. If a stable bond is formed, the GTP of the EF-Tu・GTP complex that brought the aminoacyl-tRNA is hydrolyzed to GDP.
The energy from this hydrolysis causes a conformational change in EF-Tu, and the EF-Tu・GDP complex is ejected from the ribosome.
12. Amino acid transfer and the three-base shift of the large subunit
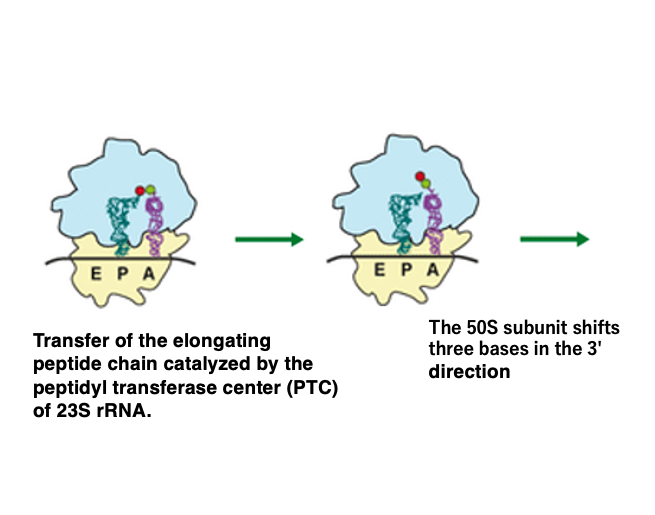
When EF-Tu・GDP dissociates from the aminoacyl tRNA, the formylmethionine attached to the initiator tRNA in the P-site is transferred to the amino acid brought by the A-site tRNA, forming a dipeptide.
At this time, only the large subunit shifts by 3 base pairs towards the 3′ direction. This amino acid transfer reaction occurs using the energy released when the aminoacyl bond is hydrolyzed. The catalyst for this reaction is the Peptidyl Transferase Center (PTC) located within the 23S rRNA in the large subunit.
13. Recycled use of EF-Tu・GTP complex
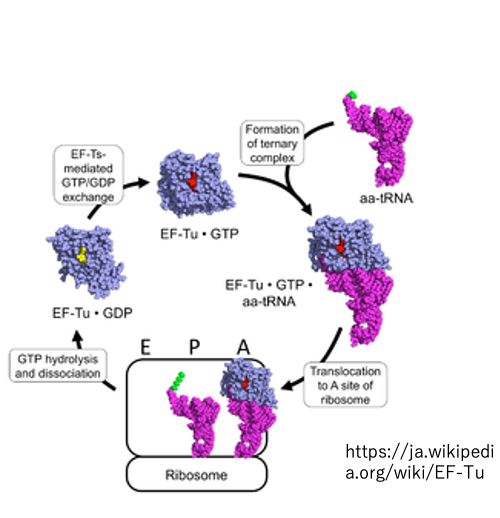
The EF-Tu・GTP complex is responsible for bringing aminoacyl-tRNA to the A-site.
After delivering the amino acid and detaching from the ribosome, the EF-Tu・GDP complex is converted back to EF-Tu・GTP for reuse. Exclusively for the initiator tRNA, it is the IF-2・GTP complex that brings it to the P-site.
14. Hybrid cavity formation in ribosome
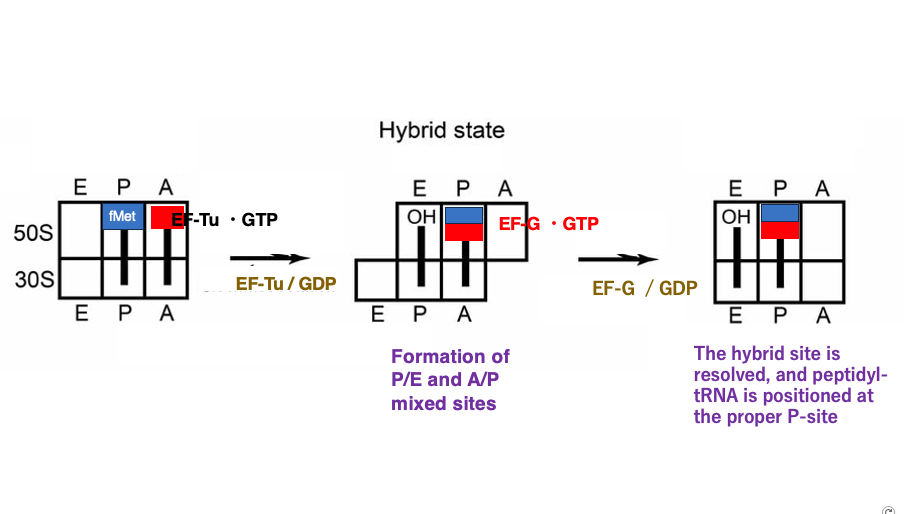
This figure shows the ribosome in a state where the large subunit has shifted 3 bases in the 3′ direction simultaneously with amino acid transfer. At this point, the E-site, P-site, and A-site of the ribosome become hybrid sites where the spatial characteristics do not match between the small and large subunits.
The initiator tRNA has already released its formylmethionine, so its 3′ end is now OH and is located in the E-site of the large subunit. However, since the small subunit has not moved, its anticodon stem remains in the P-site.
Similarly, for the tRNA carrying the dipeptide, its 3′ end aminoacyl-stem is in the P-site of the Large subunit, while its anticodon stem remains in the A-site of Small subunit.
Next, the Elongation Factor G (EF-G)•GTP complex enters the now empty A-site of the large subunit. When this GTP is hydrolyzed to GDP, the energy released causes the small subunit to shift 3 bases in the 3′ direction, resolving the hybrid sites that had formed between the large and small subunits. Once the small subunit completes its movement, the tRNA that has released its amino acid is positioned in the canonical E-site, and the tRNA with the attached dipeptide is in the canonical P-site. The initiator tRNA in the E-site will eventually be ejected from the ribosome.
15. Movement of ribosomes during peptide chain elongation
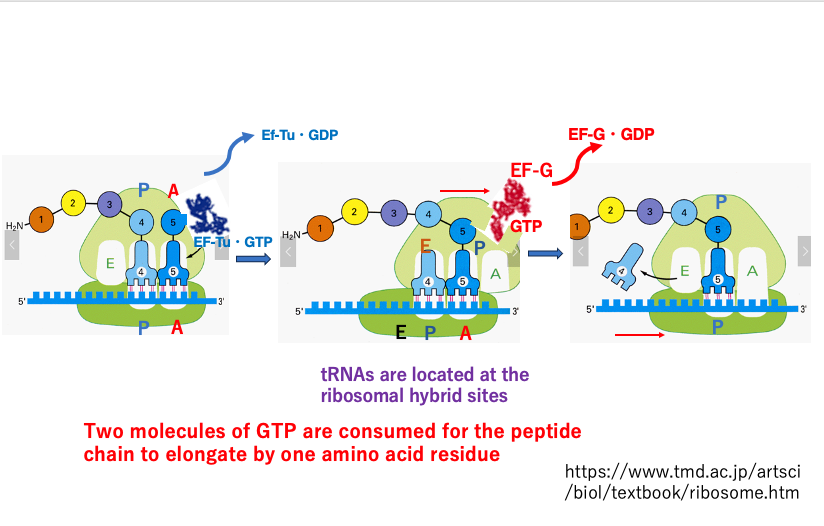
This figure illustrates a ribosome in the process of translation, with a growing peptide chain.
In the left image, an aminoacyl-tRNA and EF-Tu•GTP complex are entering the A-site. Once the codon-anticodon pairing is confirmed to be correct, GTP hydrolysis occurs, and EF-Tu•GDP dissociates from the ribosome. Simultaneously, only the large subunit shifts 3 bases in the 3′ direction, forming a hybrid site.
The now-empty A-site of the large subunit is filled by an EF-G•GTP complex (center image). The energy from the hydrolysis of this GTP causes the 30S small subunit to shift in the 3′ direction, resolving the hybrid site (right image). As a result, the peptide chain is transferred, and the empty tRNA, now positioned in the regular E-site, dissociates from the ribosome.
For the peptide chain to elongate by one amino acid residue, a total of two GTP molecules are consumed from EF-Tu•GTP and EF-G•GTP. In addition to this, the energy stored in the aminoacyl bond is also consumed.
16. Speed and Accuracy of peptide chain elongation
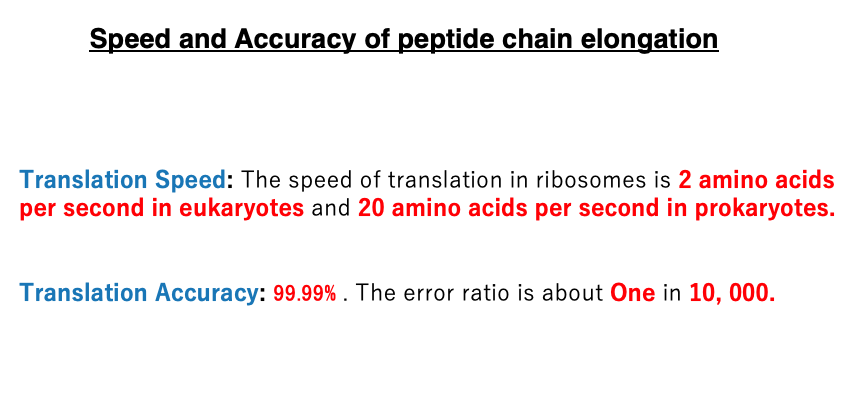
The elongation rate of peptide chains is approximately 2 amino acid residues per second in eukaryotes, while in prokaryotes, it’s much faster at about 20 amino acid residues per second.
The error rate is low, with about 1 error per 10,000 amino acids added, regardless of whether it’s a eukaryote or prokaryote. This translates to an accuracy of 99.99%
17. Mechanism of translation termination
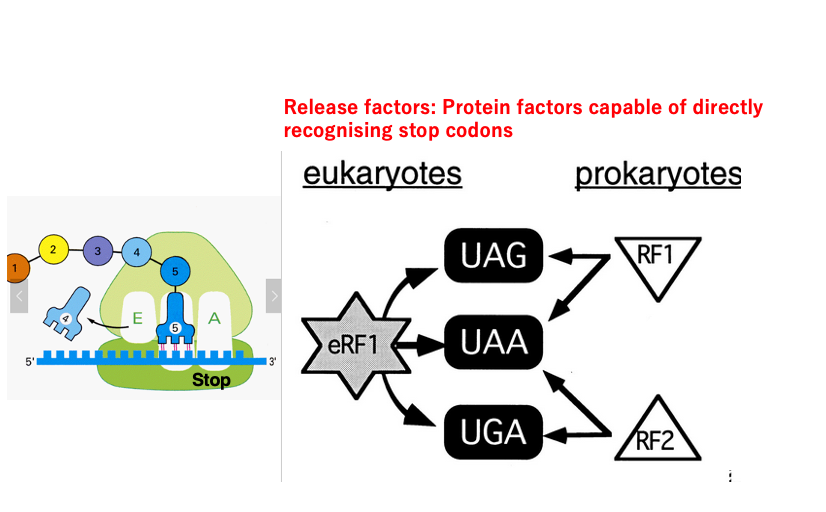
Next, I will explain the mechanism of translation termination. There are three stop codons: UAG, UAA, and UGA. However, there are no tRNAs in cells with anticodons that can form stable hydrogen bonds with these stop codons. Instead, there are protein factors called Release Factors (RF) that can bind to these stop codons. In bacteria, RF1 can recognize UAG and UAA, while RF2 can recognize UAA and UGA.
On the other hand, in eukaryotes, a Release Factor called eRF1 can bind to all three types of stop codons. Although eRF1 is also a protein molecule, its three-dimensional structure is known to be similar to that of tRNA.
In contrast, the three-dimensional structures of RF1/RF2 in eubacteria do not show similarity to tRNA. This suggests that while eRF1 and RF1/RF2 are proteins that perform the same function of translation termination, they are not considered to have evolutionary continuity.
18. Release of synthesized peptide and Dissociation of ribosome subunits
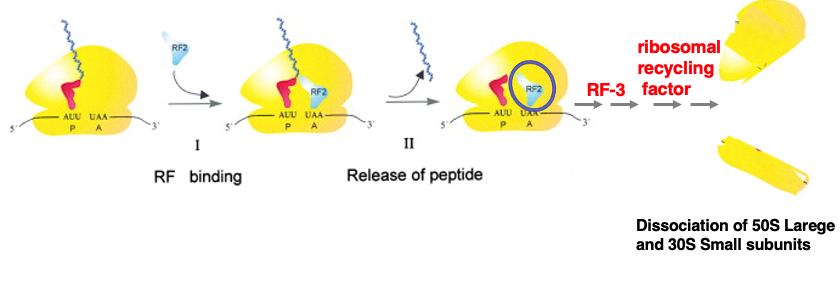
As translation progresses, a stop codon enters the A-site. At this point, the P-site contains a tRNA with an elongated nascent peptide chain attached. An appropriate Release Factor binds to the stop codon. The Release Factor then acts on the Peptidyl Transferase Center (PTC) located in the 23S rRNA of the Large subunit, catalyzing the hydrolysis of the aminoacyl bond between the nascent peptide chain and the tRNA.
The released nascent peptide chain exits through an exit tunnel in the Large subunit of the ribosome. As a result, only the deacylated tRNA and the Release Factor remain in the ribosome.
For the ribosome to initiate translation again, the empty tRNA and Release Factor must be removed, and the Large and Small subunits need to dissociate. This process involves Release Factor 3 and the ribosomal recycling factor. I have explained the mechanism of translation in bacteria.
That’s all. See you again soon.
コメント